I get the read the subfindID. But when I want to read the detailed information about it through its subfindID.
I get the subfindID from the code:
def trans(basePath,subhaloID,SnapNum,id):
'''try to get the subfindID from its subhaloID'''
RowNum, LastProgID, SubhaloID = il.sublink.treeOffsets(basePath, SnapNum, id, "SubLink")
rowStart = RowNum
rowEnd = RowNum + (LastProgID - SubhaloID)
nRows = rowEnd - rowStart + 1
offsets = il.sublink.subLinkOffsets(basePath, "SubLink", True)
rowOffsets = rowStart - offsets
fileNum = np.max(np.where(rowOffsets >= 0))
subhaloID=subhaloID%1000000
with h5py.File(il.sublink.treePath(basePath, "SubLink", fileNum), 'r') as f:
SubfindID=f['SubfindID'][subhaloID]
return SubfindID
and I want to use it in loadSingle, but it has the error:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-8-9083be1f37e1> in <module>
4 if i in cut:
5 continue
----> 6 cosI=angular(basePath,mergerlist[2*i],mergerlist[2*i+1],snap)
7 total.append(cosI)
<ipython-input-3-3e61878b4f88> in angular(basePath, SubfindID1, SubfindID2, SnapNum)
2 '''know the cos of the two galaxies' angular monmentous '''
3 sub1=il.groupcat.loadSingle(basePath, SnapNum, haloID=-1,subhaloID=SubfindID1)
----> 4 sub2=il.groupcat.loadSingle(basePath, SnapNum, haloID=-1,subhaloID=SubfindID2)
5 L1=sub1['SubhaloMass']*sub1['SubhaloSpin']
6 L2=sub2['SubhaloMass']*sub2['SubhaloSpin']
~/illustris_python/groupcat.py in loadSingle(basePath, snapNum, haloID, subhaloID)
152 with h5py.File(gcPath(basePath, snapNum, fileNum), 'r') as f:
153 for haloProp in f[gName].keys():
--> 154 result[haloProp] = f[gName][haloProp][groupOffset]
155
156 return result
h5py/_objects.pyx in h5py._objects.with_phil.wrapper()
h5py/_objects.pyx in h5py._objects.with_phil.wrapper()
/opt/conda/lib/python3.6/site-packages/h5py/_hl/dataset.py in __getitem__(self, args)
474
475 # Perform the dataspace selection.
--> 476 selection = sel.select(self.shape, args, dsid=self.id)
477
478 if selection.nselect == 0:
/opt/conda/lib/python3.6/site-packages/h5py/_hl/selections.py in select(shape, args, dsid)
92
93 sel = SimpleSelection(shape)
---> 94 sel[args]
95 return sel
96
/opt/conda/lib/python3.6/site-packages/h5py/_hl/selections.py in __getitem__(self, args)
259 return self
260
--> 261 start, count, step, scalar = _handle_simple(self.shape,args)
262
263 self._id.select_hyperslab(start, count, step)
/opt/conda/lib/python3.6/site-packages/h5py/_hl/selections.py in _handle_simple(shape, args)
449 else:
450 try:
--> 451 x,y,z = _translate_int(int(arg), length)
452 s = True
453 except TypeError:
/opt/conda/lib/python3.6/site-packages/h5py/_hl/selections.py in _translate_int(exp, length)
469
470 if not 0<=exp<length:
--> 471 raise ValueError("Index (%s) out of range (0-%s)" % (exp, length-1))
472
473 return exp, 1, 1
ValueError: Index (62467) out of range (0-14077)
so what's wrong, please help me
Dylan Nelson
14 Aug '23
I am not sure what the trans() function is trying to do?
And what does subhaloID=subhaloID%1000000 mean?
Xu Li
14 Aug '23
Dear sir,
the trans() function trys get the subfindID of the NextProgenitor which comes from tree. And for subhaloID=subhaloID%1000000 , it's used to read the index of the Nextprogenitor.
Dylan Nelson
14 Aug '23
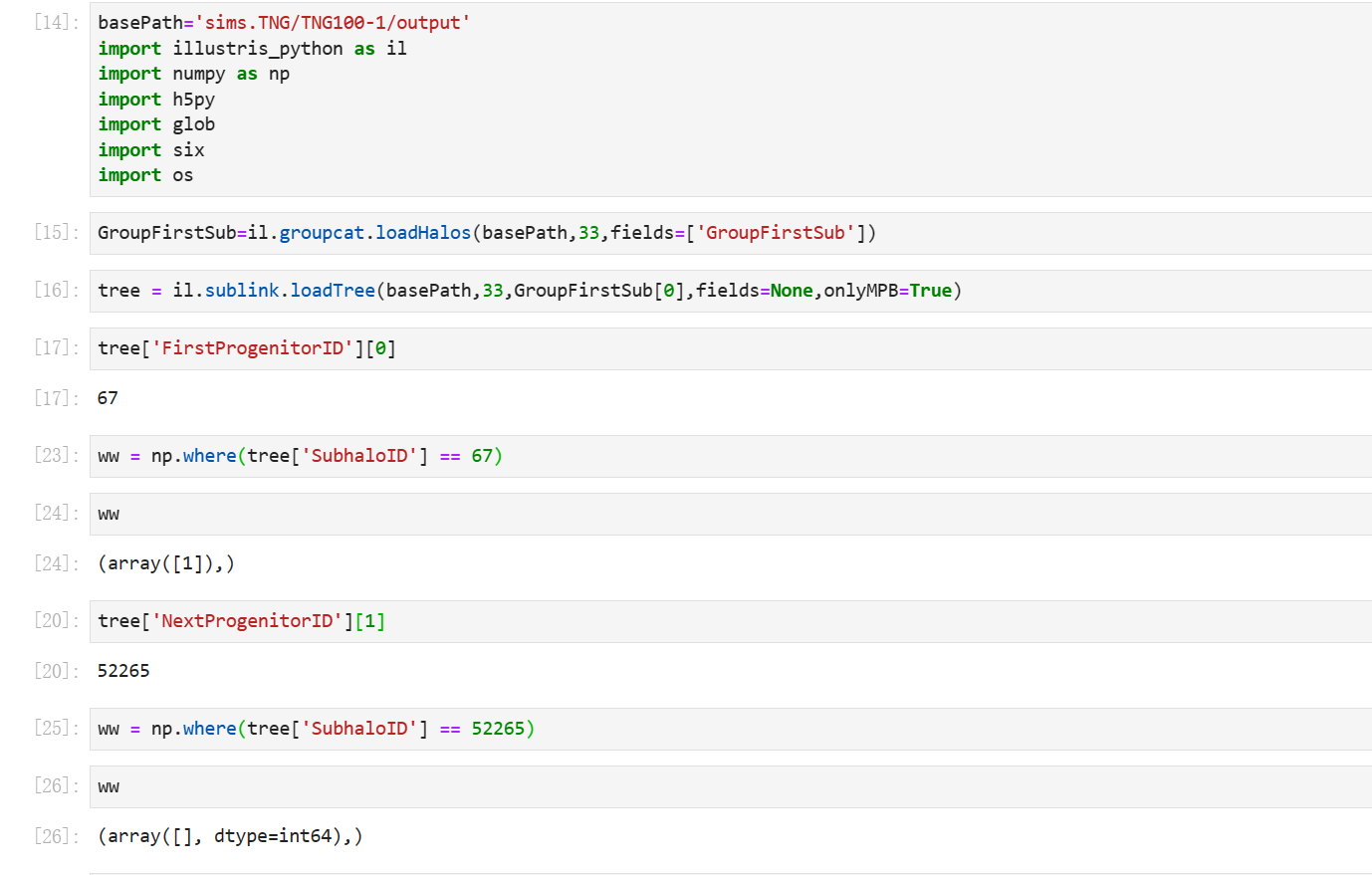
For example
In [3]: tree = il.sublink.loadTree(basePath, snap_num, subhalo_id)
In [8]: tree['FirstProgenitorID'][0]
Out[8]: 257735
In [9]: ww = np.where(tree['SubhaloID'] == 257735)
In [10]: ww
Out[10]: (array([1]),)
In [13]: tree['NextProgenitorID'][1]
Out[13]: 258657
In [14]: ww = np.where(tree['SubhaloID'] == 258657)
In [15]: ww
Out[15]: (array([923]),)
In [16]: tree['SubfindID'][923]
Out[16]: ANSWER
Xu Li
15 Aug '23
but it maybe unread because the ww of np.where(tree['SubhaloID'] == 258657) equals array[].
Dylan Nelson
15 Aug '23
By definition, if you specify onlyMPB=True when loading the tree, then the result does not contain any Next Progenitors.
If you want to explore the full tree, and not just the Main Progenitor Branch, you should delete onlyMPB=True.
Xu Li
17 Aug '23
Dear Sir,
I have a try in my code. But in 33snapshot,the array is empty.
why is the shape of the subhalo smaller than the NextProgenitorID?
I get the read the subfindID. But when I want to read the detailed information about it through its subfindID.
I get the subfindID from the code:
and I want to use it in loadSingle, but it has the error:
so what's wrong, please help me
I am not sure what the
trans()function is trying to do?And what does
subhaloID=subhaloID%1000000mean?Dear sir,
the trans() function trys get the subfindID of the NextProgenitor which comes from tree. And for subhaloID=subhaloID%1000000 , it's used to read the index of the Nextprogenitor.
For example
but it maybe unread because the ww of np.where(tree['SubhaloID'] == 258657) equals array[].
By definition, if you specify
onlyMPB=Truewhen loading the tree, then the result does not contain any Next Progenitors.If you want to explore the full tree, and not just the Main Progenitor Branch, you should delete
onlyMPB=True.Dear Sir,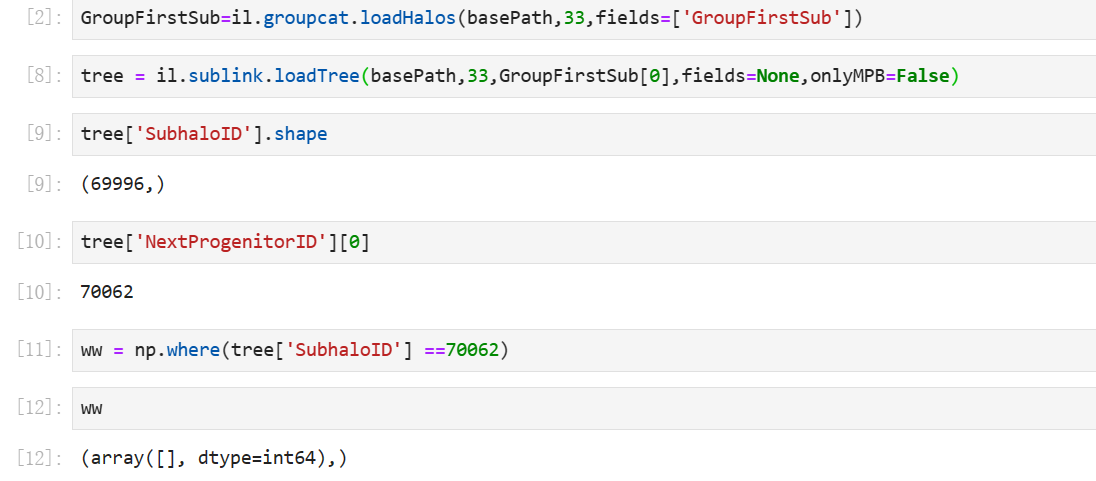
I have a try in my code. But in 33snapshot,the array is empty.
why is the shape of the subhalo smaller than the NextProgenitorID?
You cannot access
tree['NextProgenitorID'][0], this is undefined (for the root node).You should first access
tree['FirstProgenitorID'][0], which is1, thentree['NextProgenitorID'][1], which is52265.