Public Data Access Overview / Web API Documentation
The web-based interface (API) can respond to a variety of user requests and queries, and can be used in addition to, or in place of, the download and local analysis of large data files. At a high level, the API allows a user to search, extract, visualize, and analyze. In each case, the goal is to reduce the data response size, either by extracting an unmodified subset, or by calculating a derivative quantity.
This page has three sections: getting started guide, cookbook, reference.
We provide examples of accessing the API in a few languages. Select one to show all the content on this page specifically for that language.
Note! IDL v8.2+ required, for the
json_parse()
function. Otherwise, you will have to parse JSON responses yourself.
API Getting Started Guide
First, start up your interface of choice and define a helper function, whose purpose is to make a HTTP GET request to a specified URL ("endpoint"), and verify that the response is successful. If the response type is JSON, then automatically decode it into a dict-like object.
import requests
baseUrl = 'http://www.tng-project.org/api/'
headers = {"api-key":"INSERT_API_KEY_HERE"}
def get(path, params=None):
# make HTTP GET request to path
r = requests.get(path, params=params, headers=headers)
# raise exception if response code is not HTTP SUCCESS (200)
r.raise_for_status()
if r.headers['content-type'] == 'application/json':
return r.json() # parse json responses automatically
return r
IDL> baseUrl = 'http://www.tng-project.org/api/'
function get, path, params=p
headers = 'api-key: INSERT_API_KEY_HERE'
oUrl = OBJ_NEW('IDLnetUrl')
oUrl->SetProperty, headers=headers
query = ''
if n_elements(p) gt 0 then begin
if ~isa(p,'hash') then p = hash(p) ; optionally convert struct input
foreach key,p.keys() do query += strlowcase(key) + '=' + str(p[key]) + '&'
query = '?' + strmid(query,0,strlen(query)-1)
endif
r = oUrl->Get(url=path+query,/buffer)
oUrl->GetProperty, content_type=content_type
OBJ_DESTROY, oUrl
if content_type eq 'json' then r = json_parse(string(r))
return, r
end
>> baseUrl = 'http://www.tng-project.org/api/';
>>
>> function r = get_url(url,params)
>>
>> header1 = struct('name','api-key','value','INSERT_API_KEY_HERE');
>> header2 = struct('name','accept','value','application/json,octet-stream');
>> header = struct([header1,header2]);
>>
>> if exist('params','var')
>> keys = fieldnames(params);
>> query = '';
>> for i=1:numel(keys), query = strcat( query, '&', keys(i), '=', num2str(params.(keys{i})) );, end
>> url = strcat(url, '?', query);
>> url = url{1};
>> end
>>
>> [r,extras] = urlread2(url, 'GET', '', header);
>>
>> if ~extras.isGood, error(['error: ' num2str(extras.status.value) ' ' extras.status.msg]);, end
>>
>> if extras.firstHeaders.('Content_Type') == 'application/json', r = loadjson(r);, end
>>
>> end
julia> TODO
Issue a request to the API root.
>>> r = get(baseUrl)
IDL> r = get(baseUrl)
>> r = get_url(baseUrl);
julia>
The response is a dictionary object with one key, "simulations", which is a list of
N (currently 18 in this example) available runs:
>>> r.keys()
['simulations']
>>> len(r['simulations'])
18
IDL> r.keys()
[
"simulations"
]
IDL> r['simulations'].count()
18
>> r
r =
simulations: {1x18 cell}
julia>
Print out all fields of the first, as well as the names of all eighteen.
>>> r['simulations'][0]
{'name': 'Illustris-1',
'num_snapshots': 134,
'url': 'http://www.tng-project.org/api/Illustris-1/'}
>>> names = [sim['name'] for sim in r['simulations']]
>>> names
['Illustris-1',
'Illustris-1-Dark',
'Illustris-2',
'Illustris-2-Dark',
'Illustris-3',
'Illustris-3-Dark',
'Illustris-1-Subbox0',
'Illustris-1-Subbox1',
'Illustris-1-Subbox2',
'Illustris-1-Subbox3',
'Illustris-2-Subbox0',
'Illustris-2-Subbox1',
'Illustris-2-Subbox2',
'Illustris-2-Subbox3',
'Illustris-3-Subbox0',
'Illustris-3-Subbox1',
'Illustris-3-Subbox2',
'Illustris-3-Subbox3']
IDL> ( r['simulations'] )[0]
{
"name": "Illustris-1",
"num_snapshots": 134,
"url": "http://www.tng-project.org/api/Illustris-1/"
}
IDL> names = list()
IDL> foreach sim,r['simulations'] do names.add, sim['name']
IDL> names
[
"Illustris-1",
"Illustris-1-Dark",
"Illustris-2",
"Illustris-2-Dark",
"Illustris-3",
"Illustris-3-Dark",
"Illustris-1-Subbox0",
"Illustris-1-Subbox1",
"Illustris-1-Subbox2",
"Illustris-1-Subbox3",
"Illustris-2-Subbox0",
"Illustris-2-Subbox1",
"Illustris-2-Subbox2",
"Illustris-2-Subbox3",
"Illustris-3-Subbox0",
"Illustris-3-Subbox1",
"Illustris-3-Subbox2",
"Illustris-3-Subbox3"
]
>> r.('simulations'){1}
ans =
name: 'Illustris-1'
num_snapshots: 134
url: 'http://www.tng-project.org/api/Illustris-1/'
>> names = {}
>> for i=1:numel(r.('simulations')), names{i} = r.('simulations'){i}.('name');, end
>> names
names =
Columns 1 through 18
'Illustris-1' 'Illustris-1-Dark' 'Illustris-2' 'Illustris-2-Dark'
'Illustris-3' 'Illustris-3-Dark' 'Illustris-1-Subbox0' 'Illustris-1-Subbox1'
'Illustris-1-Subbox2' 'Illustris-1-Subbox3' 'Illustris-2-Subbox0'
'Illustris-2-Subbox1' 'Illustris-2-Subbox2' 'Illustris-2-Subbox3'
'Illustris-3-Subbox0' 'Illustris-3-Subbox1' 'Illustris-3-Subbox2'
'Illustris-3-Subbox3'
julia>
We see the three resolution levels of Illustris, the three dark matter only runs, and the four
subboxes per "full physics" run, as expected. Each entry has only three fields: name,
num_snapshots, and url. We can retrieve the full metadata for a particular
simulation by submitting a request to the specified url.
Additional simulations.
Note that you will now also see the three baryonic runs for TNG100, the three baryonic runs of TNG300, the six corresponding dark matter only analogs, and several subboxes for each baryonic run.
Let's look at Illustris-3 by determining which entry in r it is, then requesting
the url field of that entry.
>>> i = names.index('Illustris-3')
>>> i
4
>>> sim = get( r['simulations'][i]['url'] )
>>> sim.keys()
['softening_dm_max_phys',
'omega_0',
'snapshots',
...
'softening_dm_comoving',
'softening_gas_comoving']
>>> sim['num_dm']
94196375
IDL> i = names.where('Illustris-3')
IDL> i
4
IDL> sim = get( r['simulations',i,'url'] )
IDL> sim.keys()
[
"name",
"description",
...
"files",
"checksums",
"snapshots"
]
IDL> sim['num_dm']
94196375
>> [~,i] = ismember('Illustris-3',names)
i =
5
>> sim = get_url( r.('simulations'){i}.('url') );
>> fieldnames(sim)
ans =
'name'
'description'
...
'files'
'checksums'
'snapshots'
>> sim.('num_dm')
ans =
94196375
julia>
Notice how we do not actually need to construct the URL by hand.
This is in general true: whenever an API response refers to another resource or endpoint, it does so with an absolute URL, which can be directly followed to retrieve that resource. Meaning, that there is no need to know the structure of the API in order to navigate it.
In this case, we could have seen from the reference table at the bottom of this page, that the endpoint which
retrieves the full metadata for a given simulation is /api/{sim_name}/. Therefore, we could
manually construct the URL www.tng-project.org/api/Illustris-3/ and send a request.
Alternatively, we can simply follow the url field that we already have to arrive at the same place.
Next, get the snapshot listing for this simulation.
>>> sim['snapshots']
'http://www.tng-project.org/api/Illustris-3/snapshots/'
>>> snaps = get( sim['snapshots'] )
>>> len(snaps)
136
IDL> sim['snapshots']
http://www.tng-project.org/api/Illustris-3/snapshots/
IDL> snaps = get( sim['snapshots'] )
IDL> snaps.count()
136
>> sim.('snapshots')
ans =
http://www.tng-project.org/api/Illustris-3/snapshots/
>> snaps = get_url( sim.('snapshots') );
>> numel(snaps)
ans =
136
julia>
There are 136 total snapshots, inspect the last one, which corresponds to $z=0$.
>>> snaps[-1]
{'num_groups_subfind': 121209,
'number': 135,
'redshift': 2.2204460492503099e-16,
'url': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/'}
IDL> snaps[-1]
{
"number": 135,
"redshift": 2.2204460492503099e-16,
"num_groups_subfind": 121209,
"url": "http://www.tng-project.org/api/Illustris-3/snapshots/135/"
}
>> snaps{end}
ans =
number: 135
redshift: 2.2204e-16
num_groups_subfind: 121209
url: [1x70 char]
>> snaps{end}.('url')
ans =
http://www.tng-project.org/api/Illustris-3/snapshots/135/
julia>
Retrieve the full meta-data for this snapshot.
>>> snap = get( snaps[-1]['url'] )
>>> snap
{'files': {'groupcat': 'http://www.tng-project.org/api/Illustris-3/files/groupcat-135/',
'snapshot': 'http://www.tng-project.org/api/Illustris-3/files/snapshot-135/'},
'filesize_groupcat': 114056740.0,
'filesize_rockstar': 0.0,
'filesize_snapshot': 23437820660.0,
'num_bhs': 33582,
'num_dm': 94196375,
'num_gas': 87571556,
'num_groups_fof': 131727,
'num_groups_rockstar': 0,
'num_groups_subfind': 121209,
'num_stars': 4388167,
'num_trmc': 94196375,
'number': 135,
'redshift': 2.2204460492503099e-16,
'simulation': 'http://www.tng-project.org/api/Illustris-3/',
'subhalos': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/',
'url': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/'}
IDL> snap = get( snaps[-1,'url'] )
IDL> snap
{
"simulation": "http://www.tng-project.org/api/Illustris-3/",
"number": 135,
"redshift": 2.2204460492503099e-16,
"num_gas": 87571556,
"num_dm": 94196375,
"num_trmc": 94196375,
"num_stars": 4388167,
"num_bhs": 33582,
"num_groups_fof": 131727,
"num_groups_subfind": 121209,
"num_groups_rockstar": 0,
"filesize_snapshot": 23437820660.000000,
"filesize_groupcat": 114056740.00000000,
"filesize_rockstar": 0.0000000000000000,
"url": "http://www.tng-project.org/api/Illustris-3/snapshots/135/",
"subhalos": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/",
"files": {
"snapshot": "http://www.tng-project.org/api/Illustris-3/files/snapshot-135/",
"groupcat": "http://www.tng-project.org/api/Illustris-3/files/groupcat-135/"
}
}
>> snap = get_url( snaps{end}.('url') )
snap =
simulation: 'http://www.tng-project.org/api/Illustris-3/'
number: 135
redshift: 2.2204e-16
num_gas: 87571556
num_dm: 94196375
num_trmc: 94196375
num_stars: 4388167
num_bhs: 33582
num_groups_fof: 131727
num_groups_subfind: 121209
num_groups_rockstar: 0
filesize_snapshot: 2.3438e+10
filesize_groupcat: 114056740
filesize_rockstar: 0
url: [1x70 char]
subhalos: [1x79 char]
files: [1x1 struct]
julia>
In addition to numeric meta-data fields such as num_gas at this snapshot, we have the
url field, describing the location of this particular snapshot, which is exactly the URL
which we just requested. The simulation links back to the parent simulation which owns this
snapshot, while subhalos links deeper, to all the child subhalos which belong to this
snapshot. Finally, the files dict contains entries for all "raw file" downloads available
specifically for this snapshot. These are the same links you will find embedded in the wget
commands for snapshot 135 on the Illustris-3 Downloads
page.
Request and inspect the subhalos endpoint.
>>> subs = get( snap['subhalos'] )
>>> subs.keys()
['count', 'previous', 'results', 'next']
>>> subs['count']
121209
>>> subs['next']
'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?offset=100'
>>> len(subs['results'])
100
IDL> subs = get( snap['subhalos'] )
IDL> subs.keys()
[
"count",
"next",
"previous",
"results"
]
IDL> subs['count']
121209
IDL> subs['next']
http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?offset=100
IDL> subs['results'].count()
100
>> subs = get_url( snap.('subhalos') );
>> fieldnames(subs)
ans =
'count'
'next'
'previous'
'results'
>> subs.('count')
ans =
121209
>> subs.('next')
ans =
http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?offset=100
>> numel(subs.('results'))
ans =
100
julia>
The response is a paginated list of all Subfind subhalos which exist at this snapshot.
The default page size is 100 elements, this can be overridden by specifying a limit
parameter.
>>> subs = get( snap['subhalos'], {'limit':220} )
>>> len(subs['results'])
220
>>> subs['next']
'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?limit=220&offset=220'
>>> subs['results'][0]
{'id': 21246,
'mass_log_msun': 10.671343457957859,
'url': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/21246/'}
IDL> subs = get( snap['subhalos'], params={limit:220} )
IDL> subs['results'].count()
100
IDL> subs['next']
http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?offset=100
IDL> subs['results',0]
{
"id": 0,
"mass_log_msun": 14.557165062415164,
"url": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/0/"
}
>> subs = get_url( snap.('subhalos'), struct('limit',220) );
>> numel(subs.('results'))
ans =
220
>> subs.('next')
ans =
http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?limit=220&offset=220
>> subs.('results'){1}
ans =
id: 0
mass_log_msun: 14.5572
url: [1x81 char]
julia>
Each element of results contains minimal information: the subhalo id,
its total mass (log solar units), and its unique URL. Note that, although this is the first result
of the first page, the ID may not necessarily be zero!
Note: Ordering of subhalo searches.
Return order is arbitrary unless specified.Request the first twenty subhalos at this snapshot, sorted by descending stellar mass.
>>> subs = get( snap['subhalos'], {'limit':20, 'order_by':'-mass_stars'} )
>>> len(subs['results'])
20
>>> [ subs['results'][i]['id'] for i in range(5) ]
[0,
1030,
2074,
2302,
2843]
IDL> subs = get( snap['subhalos'], params={limit:20, order_by:'-mass_stars'} )
IDL> subs['results'].count()
20
IDL> for i=0,4 do print, subs['results',i,'id']
0
1030
2074
2302
2843
>> subs = get_url( snap.('subhalos'), struct('limit',20,'order_by','-mass_stars') );
>> numel(subs.('results'))
ans =
20
>> for i=1:5, disp( subs.('results'){i}.('id') ), end
0
1030
2074
2302
2843
julia>
Just to be clear, the full URL which was just requested was:
www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/?limit=20&order_by=-mass_stars
(if you are logged in, browsing to this location will dump you into the Browsable API).
Note: Ordering
You can order by any field in the catalog.
The negative sign indicates descending order, otherwise ascending order is assumed.
As expected, because subhalos within each halo are ordered roughly in order of decreasing total mass, the most massive subhalo (with ID==0) also has the most stars. The next ID (1030) is likely a central subhalo of a subsequent FoF halo.
Let's check. First, get the full subhalo information for ID==1030.
>>> sub = get( subs['results'][1]['url'] )
>>> sub
{'bhmdot': 0.199144,
'cm_x': 10992.8,
...
'cutouts': {'parent_halo': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/cutout.hdf5',
'subhalo': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/cutout.hdf5'},
'desc_sfid': -1,
'desc_snap': -1,
...
'grnr': 2,
...
'id': 1030,
...
'meta': {'info': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/info.json',
'simulation': 'http://www.tng-project.org/api/Illustris-3/',
'snapshot': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/',
'url': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/'},
...
'primary_flag': 1,
'prog_sfid': 1004,
'prog_snap': 134,
'related': {'parent_halo': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/',
'sublink_descendant': None,
'sublink_progenitor': 'http://www.tng-project.org/api/Illustris-3/snapshots/134/subhalos/1004/'},
...
'snap': 135,
...
'supplementary_data': {},
'trees': {'lhalotree': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/lhalotree/full.hdf5',
'lhalotree_mpb': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/lhalotree/mpb.hdf5',
'sublink': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/sublink/full.hdf5',
'sublink_mpb': 'http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/sublink/mpb.hdf5'},
...}
IDL> sub = get( subs['results',1,'url'] )
IDL> sub
{
"snap": 135,
"id": 1030,
"bhmdot": 0.19914399999999999,
...
"desc_sfid": -1,
"desc_snap": -1,
...
"grnr": 2,
...
"primary_flag": 1,
"prog_sfid": 1004,
"prog_snap": 134,
...
"supplementary_data": {},
"cutouts": {"parent_halo": "http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/cutout.hdf5",
"subhalo": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/cutout.hdf5"},
"meta": {"info": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/info.json",
"simulation": "http://www.tng-project.org/api/Illustris-3/",
"snapshot": "http://www.tng-project.org/api/Illustris-3/snapshots/135/",
"url": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/"},
"related": {"parent_halo": "http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/",
"sublink_descendant": null,
"sublink_progenitor": "http://www.tng-project.org/api/Illustris-3/snapshots/134/subhalos/1004/"},
"trees": {"lhalotree": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/lhalotree/full.hdf5",
"lhalotree_mpb": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/lhalotree/mpb.hdf5",
"sublink": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/sublink/full.hdf5",
"sublink_mpb": "http://www.tng-project.org/api/Illustris-3/snapshots/135/subhalos/1030/sublink/mpb.hdf5"},
...}
>> sub = get_url( subs.('results'){2}.('url') )
sub =
snap: 135
id: 1030
bhmdot: 0.1991
...
prog_snap: 134
prog_sfid: 1004
desc_snap: -1
desc_sfid: -1
grnr: 2
primary_flag: 1
...
related: [1x1 struct]
cutouts: [1x1 struct]
trees: [1x1 struct]
supplementary_data: [1x1 struct]
meta: [1x1 struct]
>> sub.('related')
ans =
sublink_progenitor: [1x84 char]
sublink_descendant: []
parent_halo: [1x78 char]
>> sub.('related').('sublink_progenitor')
ans =
http://www.tng-project.org/api/Illustris-3/snapshots/134/subhalos/1004/
julia>
The response is a combination of numeric fields and links to related objects, as well as additional data.
For example, desc_sfid = -1 and desc_snap = -1 indicate that this subhalo has no
descendant in the SubLink trees (as expected, since we are at $z=0$). On the other hand, prog_sfid = 1004
and prog_snap = 134 indicate that the main progenitor of this subhalo has ID 1004 at
snapshot 134. The related['sublink_progenitor'] link would take us directly there.
We also have id = 1030, a good sanity check. grnr = 2 indicates that this subhalo is a
member of FoF 2. primary_flag = 1 indicates that this is the central (i.e. most massive, or "primary")
subhalo of this FoF halo.
Let us directly request a group catalog field dump of the parent FoF halo.
>>> url = sub['related']['parent_halo'] + "info.json"
>>> url
'http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/info.json'
>>> parent_fof = get(url)
>>> parent_fof.keys()
['SnapshotNumber', 'SimulationName', 'InfoType', 'InfoID', 'Group']
>> parent_fof['Group']
{...
'GroupFirstSub': 1030,
...
'GroupNsubs': 366,
'GroupPos': [10908.2392578125, 50865.515625, 47651.7890625],
...}
IDL> url = sub['related','parent_halo'] + "info.json"
IDL> url
http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/info.json
IDL> parent_fof = get(url)
IDL> parent_fof.keys()
[
"SnapshotNumber",
"SimulationName",
"InfoType",
"InfoID",
"Group"
]
IDL> parent_fof['Group']
{
...
"GroupFirstSub": 1030,
...
"GroupNsubs": 366,
"GroupPos": [
10908.239257812500,
50865.515625000000,
47651.789062500000
],
...
}
>> url = [sub.('related').('parent_halo') 'info.json']
url =
http://www.tng-project.org/api/Illustris-3/snapshots/135/halos/2/info.json
>> parent_fof = get_url(url);
>> fieldnames(parent_fof)
ans =
'SnapshotNumber'
'SimulationName'
'InfoType'
'InfoID'
'Group'
>> parent_fof.('Group')
ans =
Group_M_Crit200: 1.7347e+04
...
GroupFirstSub: 1030
...
GroupPos: [1.0908e+04 5.0866e+04 4.7652e+04]
...
GroupNsubs: 366
...
julia>
Note: info.json endpoints
Thesubhalos/N/info.json and halos/N/info.json endpoints provide a raw
extraction from the group catalogs, so the fields are named accordingly.
In this case, we see that subhalo 1030 is indeed the central for this FoF 2, which has 366 total subhalos.
Let us return to the subhalo itself, and make some requests which return HDF5 data. First, extend our helper function so that if it recieves a binary response, it saves it to a file with the appropriate name (in the current working directory, customize as needed).
def get(path, params=None):
# make HTTP GET request to path
r = requests.get(path, params=params, headers=headers)
# raise exception if response code is not HTTP SUCCESS (200)
r.raise_for_status()
if r.headers['content-type'] == 'application/json':
return r.json() # parse json responses automatically
if 'content-disposition' in r.headers:
filename = r.headers['content-disposition'].split("filename=")[1]
with open(filename, 'wb') as f:
f.write(r.content)
return filename # return the filename string
return r
function get, path, params=p
headers = 'api-key: INSERT_API_KEY_HERE'
oUrl = OBJ_NEW('IDLnetUrl')
oUrl->SetProperty, headers=headers
query = ''
if n_elements(p) gt 0 then begin
if ~isa(p,'hash') then p = hash(p) ; optionally convert struct input
foreach key,p.keys() do query += strlowcase(key) + '=' + str(p[key]) + '&'
query = '?' + strmid(query,0,strlen(query)-1)
endif
r = oUrl->Get(url=path+query,/buffer)
oUrl->GetProperty, content_type=content_type
oUrl->GetProperty, response_header=r_header
OBJ_DESTROY, oUrl
if content_type eq 'json' then return, json_parse(string(r))
if content_type eq 'octet-stream' then begin
filename = stregex(r_header,'filename=(.*)(hdf5|fits)',/subexpr,/extract)
get_lun, lun & openw, lun, filename[1]+filename[2]
writeu, lun, r
close, lun & free_lun, lun
return, filename[1]+filename[2]
endif
return, string(r)
end
>> function r = get_url(url,params)
>>
>> header1 = struct('name','api-key','value','INSERT_API_KEY_HERE');
>> header2 = struct('name','accept','value','application/json,octet-stream');
>> header = struct([header1,header2]);
>>
>> if exist('params','var')
>> keys = fieldnames(params);
>> query = '';
>> for i=1:numel(keys), query = strcat( query, '&', keys(i), '=', num2str(params.(keys{i})) );, end
>> url = strcat(url, '?', query);
>> url = url{1};
>> end
>>
>> [r,extras] = urlread2(url, 'GET', '', header);
>>
>> if ~extras.isGood, error(['error: ' num2str(extras.status.value) ' ' extras.status.msg]);, end
>>
>> if strcmp(extras.firstHeaders.('Content_Type'),'application/json'), r = loadjson(r);, end
>>
>> if strcmp(extras.firstHeaders.('Content_Type'),'application/octet-stream')
>> filename = strsplit(extras.firstHeaders.('Content_Disposition'),'filename=');
>> f = fopen(filename{2},'w');
>> fwrite(f,r);
>> fclose(f);
>> r = filename{2}; % return filename string
>> end
>>
>> end
julia> TODO
Now, request the main progenitor branch from the SubLink merger trees of this subhalo.
>>> import h5py
>>> mpb1 = get( sub['trees']['sublink_mpb'] ) # file saved, mpb1 contains the filename
>>>
>>> f = h5py.File(mpb1,'r')
>>> print f.keys()
['DescendantID', ..., 'SnapNum', 'SubhaloNumber', 'SubhaloPos', ...]
>>> print len(f['SnapNum'])
104
>>> print f['SnapNum'][:]
[135 134 133 132 131 130 129 128 127 126 125 124 123 122 121 120 119 118
117 116 115 114 113 112 111 110 109 108 107 106 105 104 103 102 101 100
99 98 97 96 95 94 93 92 91 90 89 88 87 86 85 84 83 82
81 80 79 78 77 76 75 74 73 72 71 70 69 68 67 66 65 64
63 62 61 60 59 58 57 56 55 54 53 52 51 50 49 48 47 46
45 44 43 42 41 40 39 38 37 36 35 34 33 32]
>>> f.close()
IDL> mpb1 = get( sub['trees','sublink_mpb'] )
IDL> r = h5_parse(mpb1,/read)
IDL> tag_names(r)
_NAME
...
SNAPNUM
...
SUBHALONUMBER
...
SUBHALOPOS
...
IDL> r.SnapNum._dimensions[0]
104
IDL> r.SnapNum._data[*]
135 134 133 132 131 130 129 128 127
126 125 124 123 122 121 120 119 118
117 116 115 114 113 112 111 110 109
108 107 106 105 104 103 102 101 100
99 98 97 96 95 94 93 92 91
90 89 88 87 86 85 84 83 82
81 80 79 78 77 76 75 74 73
72 71 70 69 68 67 66 65 64
63 62 61 60 59 58 57 56 55
54 53 52 51 50 49 48 47 46
45 44 43 42 41 40 39 38 37
36 35 34 33 32
>> mpb1 = get_url( sub.('trees').('sublink_mpb') );
>> info = h5info(mpb1);
>> names = {};
>> for i=1:numel(info.('Datasets')), names{i} = info.('Datasets')(i).('Name');, end
>> names
names =
Columns 1 through 4
'DescendantID' 'FirstProgenitorID' [1x24 char] 'Group_M_Crit200'
...
>> SnapNum = h5read(mpb1,'/SnapNum/')
SnapNum =
135
134
...
32
julia>
Note: Available fields in tree returns
By default, extracting the main progenitor branch, or the full tree, for a given subhalo will return all fields. That is, in addition to telling you the Subfind IDs and snapshot numbers of all progenitors, the properties of each progenitor subhalo will also be returned.We see this subhalo was tracked back to snapshot 32 in the SubLink tree. For comparison, get the main progenitor branch from the LHaloTree.
>>> mpb2 = get( sub['trees']['lhalotree_mpb'] ) # file saved, mpb2 contains the filename
>>>
>>> with h5py.File(mpb2,'r') as f:
>>> print len(f['SnapNum'])
104
IDL> mpb2 = get( sub['trees','lhalotree_mpb'] )
IDL> r = h5_parse(mpb2,/read)
IDL> r.SnapNum._dimensions[0]
104
>> mpb2 = get_url( sub.('trees').('lhalotree_mpb') );
>> SnapNum2 = h5read(mpb2,'/SnapNum/');
>> numel(SnapNum2)
ans =
Su 104
julia>
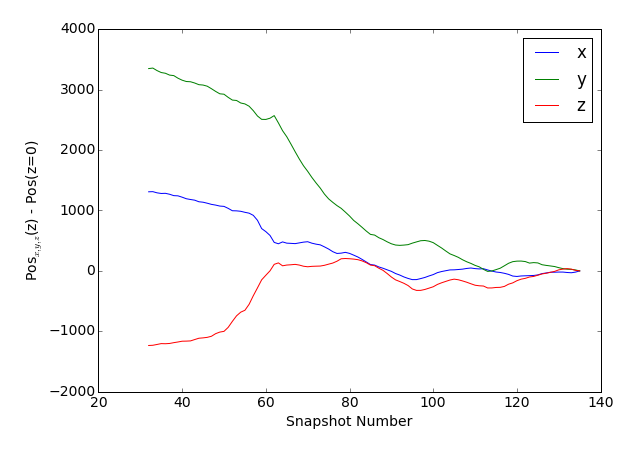
So the LHaloTree goes back to the same snapshot. Further inspection would show that in this case the tracking is similar (same $z=0$ descendant), but not identical, which is often the case. Let's plot the evolution the subhalo position, along each coordinate axis, back in time.
>>> import matplotlib.pyplot as mpl
>>> with h5py.File(mpb2,'r') as f:
>>> pos = f['SubhaloPos'][:]
>>> snapnum = f['SnapNum'][:]
>>> subid = f['SubhaloNumber'][:]
>>>
>>> for i in range(3):
>>> plt.plot(snapnum,pos[:,i] - pos[0,i], label=['x','y','z'][i])
>>> plt.legend()
>>> plt.xlabel('Snapshot Number')
>>> plt.ylabel('Pos$_{x,y,z}$(z) - Pos(z=0)');
IDL> SnapNum = r.SnapNum._data
IDL> SubhaloPos = r.SubhaloPos._data
IDL>
IDL> for i=0,2 do begin
IDL> p = plot(SnapNum, SubhaloPos[i,*]-SubhaloPos[i,0], '-', overplot=(i gt 0))
IDL> p.color = (['b','g','r'])(i)
IDL> endfor
IDL>
IDL> p.xtitle = "Snapshot Number"
IDL> p.ytitle = "Pos$_{x,y,z}$(z) - Pos(z=0)"
>> for i=1:3
>> plot(SnapNum2,SubhaloPos2(i,:)-SubhaloPos2(i,1))
>> hold on
>> end
>> legend('x','y','z');
>> xlabel('Snapshot Number');
>> ylabel('Pos$_{x,y,z}$(z) - Pos(z=0)');
julia>
Finally, let's get an actual cutout of snapshot data. Our goal: an image of the gas density around the $z=1$ progenitor of our subhalo.
First, what is the snapshot we are looking for?
>>> url = sim['snapshots'] + "z=1/"
>>> url
'http://www.tng-project.org/api/Illustris-3/snapshots/z=1/'
>>> snap = get(url)
>>> snap['number'], snap['redshift']
(85, 0.9972942257819399)
IDL> url = sim['snapshots'] + "z=1/"
IDL> url
http://www.tng-project.org/api/Illustris-3/snapshots/z=1/
IDL> snap = get(url)
IDL> snap['number'], snap['redshift']
85
0.99729422578193994
>> url = [sim.('snapshots') 'z=1/']
url =
http://www.tng-project.org/api/Illustris-3/snapshots/z=1/
>> snap = get_url(url);
>> snap.('number'), snap.('redshift')
ans =
85
ans =
0.9973
julia>
Find the target Subfind ID at snapshot 85 using the Sublink tree.
>>> i = np.where(snapnum == 85)
>>> subid[i]
185
IDL> w = where(x.SnapNum._data eq 85)
IDL> x.SubhaloNumber._data[w]
185
>> i = find(SnapNum2 == 85);
>> SubhaloNumber = h5read(mpb2,'/SubhaloNumber/');
>> SubhaloNumber(i)
ans =
185
julia>
Request the subhalo details, and a snapshot cutout consisting only of Gas fields
Coordinates,Masses.
>>> sub_prog_url = "http://www.tng-project.org/api/Illustris-3/snapshots/85/subhalos/185/"
>>> sub_prog = get(sub_prog_url)
>>> sub_prog['pos_x'], sub_prog['pos_y']
(11013.3, 51469.6)
>>> cutout_request = {'gas':'Coordinates,Masses'}
>>> cutout = get(sub_prog_url+"cutout.hdf5", cutout_request)
IDL> sub_prog_url = "http://www.tng-project.org/api/Illustris-3/snapshots/85/subhalos/185/"
IDL> sub_prog = get(sub_prog_url)
IDL> sub_prog['pos_x'], sub_prog['pos_y']
11013.299999999999
51469.599999999999
IDL> cutout_request = {gas:'Coordinates,Masses'}
IDL> cutout = get(sub_prog_url + "cutout.hdf5", params=cutout_request)
>> sub_prog_url = 'http://www.tng-project.org/api/Illustris-3/snapshots/85/subhalos/185/';
>> sub_prog = get_url(sub_prog_url);
>> sub_prog.('pos_x'), sub_prog.('pos_y')
ans =
1.1013e+04
ans =
5.1470e+04
>> cutout_request = struct('gas','Coordinates,Masses');
>> cutout = get_url( [sub_prog_url 'cutout.hdf5'], cutout_request);
julia>
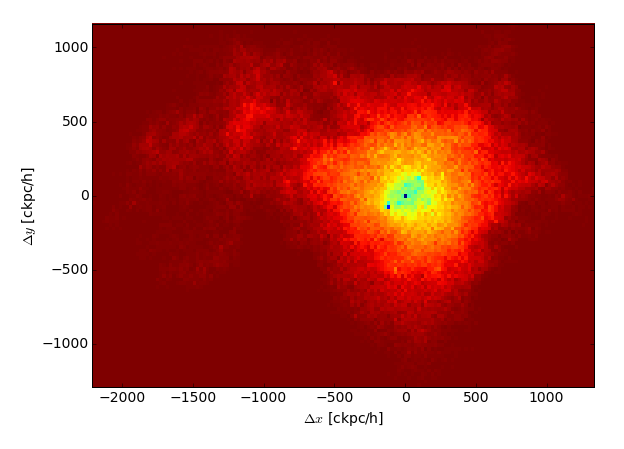
Make a quick 2d histogram visualization of the distribution of gas bound to this subhalo, weighted by the log of gas cell mass, and with position relative to the subhalo center.
>>> with h5py.File(cutout,'r') as f:
>>> x = f['PartType0']['Coordinates'][:,0] - sub_prog['pos_x']
>>> y = f['PartType0']['Coordinates'][:,1] - sub_prog['pos_y']
>>> dens = np.log10(f['PartType0']['Masses'][:])
>>>
>>> plt.hist2d(x,y,weights=dens,bins=[150,100])
>>> plt.xlabel('$\Delta x$ [ckpc/h]')
>>> plt.ylabel('$\Delta y$ [ckpc/h]');
IDL> x = r.PartType0.Coordinates._data[0,*] - sub_prog['pos_x']
IDL> y = r.PartType0.Coordinates._data[1,*] - sub_prog['pos_y']
IDL> dens = alog10( r.PartType0.Masses._data )
IDL> h2d = hist_2d(x,y, bin1=20, bin2=20) ; use hist2d or hist_nd_weight for mass-weighting
IDL> g = image(bytscl(h2d), RGB_TABLE=13, axis_style=2, $
IDL> image_dim=[max(x)-min(x),max(y)-min(y)], image_loc=[min(x), min(y)], $
IDL> xtitle='$\Delta x$ [ckpc/h]',ytitle='$\Delta x$ [ckpc/h]')
>> Coordinates = h5read(cutout,'/PartType0/Coordinates/');
>> x = Coordinates(1,:) - sub_prog.('pos_x');
>> y = Coordinates(2,:) - sub_prog.('pos_y');
>> imagesc( [min(x) max(x)], [min(y) max(y)], log10(h));
>> xlabel('$\Delta x$ [ckpc/h]');
>> ylabel('$\Delta x$ [ckpc/h]');
julia>
That's enough! You can also explore the independent examples below.
API Cookbook
Examples for how to accomplish specific tasks, covering some different API response formats.
Each example is independent, although we will use this helper function to reduce verbosity:
>>> def get(path, params=None):
>>> # make HTTP GET request to path
>>> headers = {"api-key":"INSERT_API_KEY_HERE"}
>>> r = requests.get(path, params=params, headers=headers)
>>>
>>> # raise exception if response code is not HTTP SUCCESS (200)
>>> r.raise_for_status()
>>>
>>> if r.headers['content-type'] == 'application/json':
>>> return r.json() # parse json responses automatically
>>>
>>> if 'content-disposition' in r.headers:
>>> filename = r.headers['content-disposition'].split("filename=")[1]
>>> with open(filename, 'wb') as f:
>>> f.write(r.content)
>>> return filename # return the filename string
>>>
>>> return r
function get, path, params=p
headers = 'api-key: INSERT_API_KEY_HERE'
oUrl = OBJ_NEW('IDLnetUrl')
oUrl->SetProperty, headers=headers
query = ''
if n_elements(p) gt 0 then begin
if ~isa(p,'hash') then p = hash(p) ; optionally convert struct input
foreach key,p.keys() do query += strlowcase(key) + '=' + str(p[key]) + '&'
query = '?' + strmid(query,0,strlen(query)-1)
endif
r = oUrl->Get(url=path+query,/buffer)
oUrl->GetProperty, content_type=content_type
oUrl->GetProperty, response_header=r_header
OBJ_DESTROY, oUrl
if content_type eq 'json' then return, json_parse(string(r))
if content_type eq 'octet-stream' then begin
filename = stregex(r_header,'filename=(.*)(hdf5|fits)',/subexpr,/extract)
get_lun, lun & openw, lun, filename[1]+filename[2]
writeu, lun, r
close, lun & free_lun, lun
return, filename[1]+filename[2]
endif
if content_type eq 'png' then return, r ; raw bytes
return, string(r)
end
function str, tt
return, strcompress(string(tt),/remove_all)
end
>> function r = get_url(url,params)
>>
>> header1 = struct('name','api-key','value','INSERT_API_KEY_HERE');
>> header2 = struct('name','accept','value','application/json,octet-stream');
>> header = struct([header1,header2]);
>>
>> if exist('params','var')
>> keys = fieldnames(params);
>> query = '';
>> for i=1:numel(keys), query = strcat( query, '&', keys(i), '=', num2str(params.(keys{i})) );, end
>> url = strcat(url, '?', query);
>> url = url{1};
>> end
>>
>> [r,extras] = urlread2(url, 'GET', '', header);
>>
>> if ~extras.isGood, error(['error: ' num2str(extras.status.value) ' ' extras.status.msg]);, end
>>
>> if strcmp(extras.firstHeaders.('Content_Type'),'application/json'), r = loadjson(r);, end
>>
>> if strcmp(extras.firstHeaders.('Content_Type'),'application/octet-stream')
>> filename = strsplit(extras.firstHeaders.('Content_Disposition'),'filename=');
>> f = fopen(filename{2},'w');
>> fwrite(f,r);
>> fclose(f);
>> r = filename{2}; % return filename string
>> end
>>
>> end
julia> TODO
Task 1: for Illustris-1 at $z=0$, get all the fields available for the subhalo with id=0 and print its total mass and stellar half mass radius.
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/0/"
>>> r = get(url)
>>> r['mass']
22174.8
>>> r['halfmassrad_stars']
72.0388
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/0/"
IDL> .r test3
IDL> r = get(url)
IDL> r['mass']
22174.799999999999
IDL> r['halfmassrad_stars']
72.038799999999995
>> url = 'http://www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/0/';
>> r = get_url(url);
>> r.('mass')
ans =
2.2175e+04
>> r.('halfmassrad_stars')
ans =
72.0388
julia>
Task 2: for Illustris-1 at $z=2$, search for all subhalos with total mass $10^{11.9} M_\odot < M < 10^{12.1} M_\odot$, print the number returned, and the Subfind IDs of the first five results (arbitrarily ordered, you may get different ids).
>>> # first convert log solar masses into group catalog units
>>> mass_min = 10**11.9 / 1e10 * 0.704
>>> mass_max = 10**12.1 / 1e10 * 0.704
>>> # form the search_query string by hand for once
>>> search_query = "?mass__gt=" + str(mass_min) + "&mass__lt=" + str(mass_max)
>>> search_query
'?mass__gt=55.9207077246&mass__lt=88.6283489903'
>>> # form the url and make the request
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + search_query
>>> subhalos = get(url)
>>> subhalos['count']
550
>>> ids = [ subhalos['results'][i]['id'] for i in range(5) ]
>>> ids
[109974, 110822, 123175, 107743, 95711]
IDL> mass_min = 10^11.9 / 1e10 * 0.704
IDL> mass_max = 10^12.1 / 1e10 * 0.704
IDL> ; form the search_query string by hand for once
IDL> search_query = "?mass__gt=" + str(mass_min) + "&mass__lt=" + str(mass_max)
IDL> search_query
?mass__gt=55.9207&mass__lt=88.6284
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + search_query
IDL> subhalos = get(url)
IDL> subhalos['count']
550
IDL> ids = list()
IDL> for i=0,4 do ids.add, (subhalos['results',i])['id']
IDL> ids
[
1,
1352,
5525,
6574,
12718
]
>> mass_min = 10^11.9 / 1e10 * 0.704;
>> mass_max = 10^12.1 / 1e10 * 0.704;
>>
>> % form the search_query string by hand for once
>> search_query = ['?mass__gt=' num2str(mass_min) '&mass__lt=' num2str(mass_max)]
search_query =
?mass__gt=55.9207&mass__lt=88.6283
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/' search_query];
>> subhalos = get_url(url);
>> subhalos.('count')
ans =
550
>> ids = [];
>> for i=1:5, ids(i) = subhalos.('results'){i}.('id');, end
>> ids
ids =
1 1352 5525 6574 12718
julia>
Task 3: for Illustris-1 at $z=2$, retrieve all fields for five specific Subfind IDs (from above: 109974, 110822, 123175, 107743, 95711), print the stellar mass and number of star particles in each.
>>> ids = [109974, 110822, 123175, 107743, 95711]
>>> for id in ids:
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + str(id)
>>> subhalo = get(url)
>>> print id, subhalo['mass_stars'], subhalo['len_stars']
109974 0.283605 7270
110822 0.41813 5820
123175 0.529888 11362
107743 0.648827 10038
95711 0.623781 12722
IDL> ids = [109974, 110822, 123175, 107743, 95711]
IDL>
IDL> foreach id,ids do begin
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + str(id)
IDL> subhalo = get(url)
IDL> print, id, subhalo['mass_stars'], subhalo['len_stars']
IDL> endforeach
109974 0.28360500 7270
110822 0.41813000 5820
123175 0.52988800 11362
107743 0.64882700 10038
95711 0.62378100 12722
>> ids = [109974, 110822, 123175, 107743, 95711];
>>
>> for i=1:numel(ids)
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/' num2str(ids(i))];
>> subhalo = get_url(url);
>> fprintf('%d %g %g\n', ids(i), subhalo.('mass_stars'), subhalo.('len_stars'));
>> end
109974 0.283605 7270
110822 0.41813 5820
123175 0.529888 11362
107743 0.648827 10038
95711 0.623781 12722
julia>
Task 4: for Illustris-1 at $z=2$, for five specific Subfind IDs (from above: 109974, 110822, 123175, 107743, 95711), extract and save full cutouts from the snapshot (HDF5 format).
>>> ids = [109974, 110822, 123175, 107743, 95711]
>>>
>>> for id in ids:
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + str(id) + "/cutout.hdf5"
>>> saved_filename = get(url)
>>> print id, saved_filename
109974 cutout_109974.hdf5
110822 cutout_110822.hdf5
123175 cutout_123175.hdf5
107743 cutout_107743.hdf5
95711 cutout_95711.hdf5
IDL> ids = [109974, 110822, 123175, 107743, 95711]
IDL>
IDL> foreach id,ids do begin
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + str(id) + "/cutout.hdf5"
IDL> saved_filename = get(url)
IDL> print, id + ' ' + saved_filename
IDL> endforeach
109974 cutout_109974.hdf5
110822 cutout_110822.hdf5
123175 cutout_123175.hdf5
107743 cutout_107743.hdf5
95711 cutout_95711.hdf5
>> ids = [109974, 110822, 123175, 107743, 95711];
>>
>> for i=1:numel(ids)
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/' num2str(ids(i)) '/cutout.hdf5'];
>> saved_filename = get_url(url);
>> disp([num2str(ids(i)) ' ' saved_filename]);
>> end
109974 cutout_109974.hdf5
110822 cutout_110822.hdf5
123175 cutout_123175.hdf5
107743 cutout_107743.hdf5
95711 cutout_95711.hdf5
julia>
Task 5: for Illustris-1 at $z=2$, for five specific Subfind IDs (from above: 109974, 110822, 123175, 107743, 95711), extract and save only star particles from the parent FoF halo of each subhalo.
>>> ids = [109974, 110822, 123175, 107743, 95711]
>>> params = {'stars':'all'}
>>>
>>> for id in ids:
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + str(id)
>>> sub = get(url)
>>> saved_filename = get(sub['cutouts']['parent_halo'],params)
>>> print sub['id'], sub['grnr'], saved_filename
109974 745 cutout_745.hdf5
110822 758 cutout_758.hdf5
123175 971 cutout_971.hdf5
107743 711 cutout_711.hdf5
95711 548 cutout_548.hdf5
IDL> ids = [109974, 110822, 123175, 107743, 95711]
IDL> params = {stars:'all'}
IDL>
IDL> foreach id,ids do begin
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/" + str(id)
IDL> sub = get(url)
IDL> saved_filename = get(sub['cutouts','parent_halo'],params=params)
IDL> print, sub['id'], sub['grnr'], ' ', saved_filename
IDL> endforeach
109974 745 cutout_745.hdf5
110822 758 cutout_758.hdf5
123175 971 cutout_971.hdf5
107743 711 cutout_711.hdf5
95711 548 cutout_548.hdf5
>> ids = [109974, 110822, 123175, 107743, 95711];
>> params = struct('stars','all');
>>
>> for i=1:numel(ids)
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/z=2/subhalos/' num2str(ids(i))];
>> sub = get_url(url);
>> saved_filename = get_url( sub.('cutouts').('parent_halo'), params );
>> fprintf('%d %d %s\n', sub.('id'), sub.('grnr'), saved_filename);
>> end
109974 745 cutout_745.hdf5
110822 758 cutout_758.hdf5
123175 971 cutout_971.hdf5
107743 711 cutout_711.hdf5
95711 548 cutout_548.hdf5
julia>
Task 6: for Illustris-1 at $z=2$ for Subfind ID 109974, get a cutout including only the positions and metallicities of stars, and calculate the mean stellar metallicity in solar units within the annuli $3 \rm{kpc} < r < 5 \rm{kpc}$ (proper) centered on the fiducial subhalo position.
>>> import h5py
>>> import numpy as np
>>>
>>> id = 109974
>>> redshift = 2.0
>>> params = {'stars':'Coordinates,GFM_Metallicity'}
>>>
>>> scale_factor = 1.0 / (1+redshift)
>>> little_h = 0.704
>>> solar_Z = 0.0127
>>>
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=" + str(redshift) + "/subhalos/" + str(id)
>>> sub = get(url) # get json response of subhalo properties
>>> saved_filename = get(url + "/cutout.hdf5",params) # get and save HDF5 cutout file
>>>
>>> with h5py.File(saved_filename) as f:
>>> # NOTE! If the subhalo is near the edge of the box, you must take the periodic boundary into account! (we ignore it here)
>>> dx = f['PartType4']['Coordinates'][:,0] - sub['pos_x']
>>> dy = f['PartType4']['Coordinates'][:,1] - sub['pos_y']
>>> dz = f['PartType4']['Coordinates'][:,2] - sub['pos_z']
>>> metals = f['PartType4']['GFM_Metallicity'][:]
>>>
>>> rr = np.sqrt(dx**2 + dy**2 + dz**2)
>>> rr *= scale_factor/little_h # ckpc/h -> physical kpc
>>>
>>> w = np.where( (rr >= 3.0) & (rr < 5.0) )
>>> print np.mean( metals[w] ) / solar_Z
0.248392603881
IDL> id = 109974
IDL> redshift = 2.0
IDL> params = {stars:'Coordinates,GFM_Metallicity'}
IDL>
IDL> scale_factor = 1.0 / (1+redshift)
IDL> little_h = 0.704
IDL> solar_Z = 0.0127
IDL>
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/z=" + str(redshift) + "/subhalos/" + str(id)
IDL> sub = get(url) ; get json response of subhalo properties
IDL> saved_filename = get(url + "/cutout.hdf5",params=params) ; get and save HDF5 cutout file
IDL>
IDL> f = h5_parse(saved_filename,/read)
IDL>
IDL> dx = f.PartType4.Coordinates._data[0,*] - sub['pos_x']
IDL> dy = f.PartType4.Coordinates._data[1,*] - sub['pos_y']
IDL> dz = f.PartType4.Coordinates._data[2,*] - sub['pos_z']
IDL> metals = f.PartType4.GFM_Metallicity._data
IDL>
IDL> rr = sqrt(dx^2 + dy^2 + dz^2)
IDL> rr *= scale_factor/little_h ; ckpc/h -> physical kpc
IDL>
IDL> w = where( rr ge 3.0 and rr lt 5.0 )
IDL> print, mean( metals[w] ) / solar_Z
0.248393
>> id = 109974;
>> redshift = 2.0;
>> params = struct('stars','Coordinates,GFM_Metallicity');
>>
>> scale_factor = 1.0 / (1+redshift);
>> little_h = 0.704;
>> solar_Z = 0.0127;
>>
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/z=' num2str(redshift) '/subhalos/' num2str(id)];
>> sub = get_url(url); % get json response of subhalo properties
>> saved_filename = get_url([url '/cutout.hdf5'], params); % get and save HDF5 cutout file
>>
>> Coordinates = h5read(saved_filename,'/PartType4/Coordinates/');
>> Metals = h5read(saved_filename,'/PartType4/GFM_Metallicity/');
>>
>> dx = Coordinates(1,:) - sub.('pos_x');
>> dy = Coordinates(2,:) - sub.('pos_y');
>> dz = Coordinates(3,:) - sub.('pos_z');
>> rr = sqrt(dx.^2 + dy.^2 + dz.^2);
>> rr = rr .* scale_factor/little_h; % ckpc/h -> physical kpc
>>
>> w = find( rr >= 3.0 & rr < 5.0 );
>> mean( Metals(w) ) ./ solar_Z
ans =
0.2484
julia>
Task 8: for Illustris-1 at $z=2$, for five specific Subfind IDs (from above: 109974, 110822, 123175, 107743, 95711), locate the $z=0$ descendant of each by using the API to walk down the SubLink descendant links.
>>> ids = [109974, 110822, 123175, 107743, 95711]
>>> z0_descendant_ids = [-1]*len(ids)
>>>
>>> for i,id in enumerate(ids):
>>> start_url = "http://www.tng-project.org/api/Illustris-1/snapshots/68/subhalos/" + str(id)
>>> sub = get(start_url)
>>>
>>> while sub['desc_sfid'] != -1:
>>> # request the full subhalo details of the descendant by following the sublink URL
>>> sub = get(sub['related']['sublink_descendant'])
>>> if sub['snap'] == 135:
>>> z0_descendant_ids[i] = sub['id']
>>>
>>> if z0_descendant_ids[i] >= 0:
>>> print 'Descendant of ' + str(id) + ' at z=0 is ' + str(z0_descendant_ids[i])
>>> else:
>>> print 'Descendant of ' + str(id) + ' not followed to z=0!'
Descendant of 109974 at z=0 is 41092
Descendant of 110822 at z=0 is 338375
Descendant of 123175 at z=0 is 257378
Descendant of 107743 at z=0 is 110568
Descendant of 95711 at z=0 is 260067
IDL> ids = [109974, 110822, 123175, 107743, 95711]
IDL> z0_descendant_ids = lonarr(n_elements(ids)) - 1
IDL>
IDL> for i=0,n_elements(ids)-1 do begin
IDL> start_url = "http://www.tng-project.org/api/Illustris-1/snapshots/68/subhalos/" + str(ids[i])
IDL> sub = get(start_url)
IDL>
IDL> while sub['desc_sfid'] ne -1 do begin
IDL> ; request the full subhalo details of the descendant by following the sublink URL
IDL> sub = get(sub['related','sublink_descendant'])
IDL> if sub['snap'] eq 135 then $
IDL> z0_descendant_ids[i] = sub['id']
IDL> endwhile
IDL> if z0_descendant_ids[i] ge 0 then begin
IDL> print, 'Descendant of ' + str(ids[i]) + ' at z=0 is ' + str(z0_descendant_ids[i])
IDL> endif else begin
IDL> print, 'Descendant of ' + str(ids[i]) + ' not followed to z=0!'
IDL> endelse
IDL> endfor
Descendant of 109974 at z=0 is 41092
Descendant of 110822 at z=0 is 338375
Descendant of 123175 at z=0 is 257378
Descendant of 107743 at z=0 is 110568
Descendant of 95711 at z=0 is 260067
>> ids = [109974, 110822, 123175, 107743, 95711];
>> z0_descendant_ids = zeros(numel(ids)) - 1;
>>
>> for i=1:numel(ids)
>> start_url = ['http://www.tng-project.org/api/Illustris-1/snapshots/68/subhalos/' num2str(ids(i))];
>> sub = get_url(start_url);
>>
>> while sub.('desc_sfid') ~= -1
>> % request the full subhalo details of the descendant by following the sublink URL
>> sub = get_url( sub.('related').('sublink_descendant') );
>> if sub.('snap') == 135, z0_descendant_ids(i) = sub.('id');, end
>> end
>>
>> if z0_descendant_ids(i) >= 0
>> fprintf('Descendant of %d at z=0 is %d\n', ids(i), z0_descendant_ids(i));
>> else
>> fprintf('Descendant of %d not followed to z=0!\n', ids(i));
>> end
>> end
Descendant of 109974 at z=0 is 41092
Descendant of 110822 at z=0 is 338375
Descendant of 123175 at z=0 is 257378
Descendant of 107743 at z=0 is 110568
Descendant of 95711 at z=0 is 260067
julia>
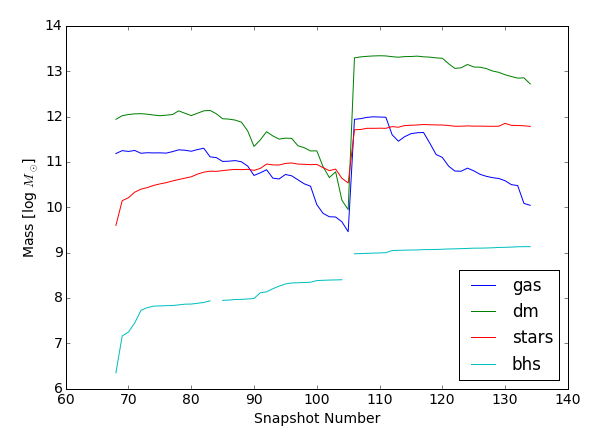
Task 9: for Illustris-1 at $z=2$ track Subfind ID 109974 to $z=0$, using the API to walk down the SubLink descendant links, and plot the mass evolution of each component (gas, dark matter, stars, and black holes).
>>> id = 109974
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/68/subhalos/" + str(id)
>>> sub = get(url) # get json response of subhalo properties
>>>
>>> # prepare dict to hold result arrays
>>> fields = ['snap','id','mass_gas','mass_stars','mass_dm','mass_bhs']
>>> r = {}
>>> for field in fields:
>>> r[field] = []
>>>
>>> while sub['desc_sfid'] != -1:
>>> for field in fields:
>>> r[field].append(sub[field])
>>> # request the full subhalo details of the descendant by following the sublink URL
>>> sub = get(sub['related']['sublink_descendant'])
>>>
>>> # make a plot (notice our subhalo falls into a much more massive halo around snapshot 105)
>>> for partType in ['gas','dm','stars','bhs']:
>>> mass_logmsun = np.log10( np.array(r['mass_'+partType])*1e10/0.704)
>>> plt.plot(r['snap'],mass_logmsun,label=partType)
>>>
>>> plt.xlabel('Snapshot Number')
>>> plt.ylabel('Mass [log $M_\odot$]')
>>> plt.legend(loc='lower right');
IDL> id = 109974
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/68/subhalos/" + str(id)
IDL> sub = get(url) ; get json response of subhalo properties
IDL>
IDL> ; prepare dict to hold result arrays
IDL> fields = ['snap','id','mass_gas','mass_stars','mass_dm','mass_bhs']
IDL> r = hash()
IDL> foreach field,fields do r[field] = list()
IDL>
IDL> while sub['desc_sfid'] ne -1 do begin
IDL> foreach field,fields do r[field].add, sub[field]
IDL> ; request the full subhalo details of the descendant by following the sublink URL
IDL> sub = get(sub['related','sublink_descendant'])
IDL> endwhile
IDL>
IDL> ; make a plot (notice our subhalo falls into a much more massive halo around snapshot 105)
IDL> foreach partType,['gas','dm','stars','bhs'],i do begin
IDL> mass_logmsun = alog10( r['mass_'+partType].toArray()*1e10/0.704 )
IDL> p = plot(r['snap'].toArray(), mass_logmsun, name=partType, overplot=(i ne 0))
IDL> p.color = (['b','g','r','m'])(i)
IDL> endforeach
IDL>
IDL> p.xtitle = 'Snapshot Number'
IDL> p.ytitle = 'Mass [log $M_\odot$]'
IDL> g = legend(target=p)
>> id = 109974;
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/68/subhalos/' num2str(id)];
>> sub = get_url(url); % get json response of subhalo properties
>>
>> % prepare struct to hold result arrays
>> fields = {'snap','id','mass_gas','mass_stars','mass_dm','mass_bhs'};
>> for i=1:numel(fields), r.(fields{i}) = [];, end
>>
>> while sub.('desc_sfid') ~= -1
>> for i=1:numel(fields), r.(fields{i}) = [ r.(fields{i}) sub.(fields{i}) ];, end
>> % request the full subhalo details of the descendant by following the sublink URL
>> sub = get_url( sub.('related').('sublink_descendant') );
>> end
>>
>> % make a plot (notice our subhalo falls into a much more massive halo around snapshot 105)
>> for i=1:numel(partTypes)
>> mass_logmsun = log10( r.(['mass_' partTypes{i}]) * 1e10/0.704 );
>> plot(r.('snap'), mass_logmsun);
>> hold all
>> end
>>
>> xlabel('Snapshot Number');
>> ylabel('Mass [log $M_\odot$]');
>> legend(partTypes,'Location','best');
julia>
Task 10: for Illustris-1 at $z=0$, check if pre-rendered mock stellar images exist for five specific Subfind IDs (the descendants from above: 41092, 338375, 257378, 110568, 260067). If so, download and display the PNGs.
>>> import matplotlib.image as mpimg
>>> from io import BytesIO
>>>
>>> ids = [41092,338375,257378,110568,260067]
>>>
>>> sub_count = 1
>>> plt.figure(figsize=[15,3])
>>>
>>> for id in ids:
>>> url = "http://www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/" + str(id)
>>> sub = get(url)
>>>
>>> # it is of course possible this data product does not exist for all requested subhalos
>>> if 'stellar_mocks' in sub['supplementary_data']:
>>> # download PNG image, the version which includes all stars in the FoF halo (try replacing 'fof' with 'gz')
>>> png_url = sub['supplementary_data']['stellar_mocks']['image_fof']
>>> response = get(png_url)
>>>
>>> # make plot a bit nicer
>>> plt.subplot(1,len(ids),sub_count)
>>> plt.text(0,-20,"ID="+str(id),color='blue')
>>> plt.gca().axes.get_xaxis().set_ticks([])
>>> plt.gca().axes.get_yaxis().set_ticks([])
>>> sub_count += 1
>>>
>>> # plot the PNG binary data directly, without actually saving a .png file
>>> file_object = BytesIO(response.content)
>>> plt.imshow(mpimg.imread(file_object))
IDL> ids = [41092,338375,257378,110568,260067]
IDL>
IDL> foreach id,ids,sub_count do begin
IDL> url = "http://www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/" + str(id)
IDL> sub = get(url)
IDL>
IDL> ; it is of course possible this data product does not exist for all requested subhalos
IDL> if ~sub['supplementary_data'].hasKey('stellar_mocks') then continue
IDL>
IDL> ; download PNG image, the version which includes all stars in the FoF halo (try replacing 'fof' with 'gz')
IDL> png_url = sub['supplementary_data','stellar_mocks','image_fof']
IDL> response = get(png_url)
IDL>
IDL> ; save PNG to temporary file and read it (cannot easily decode PNG format in-memory)
IDL> get_lun, lun & openw, lun, 'out.png'
IDL> writeu, lun, response
IDL> close, lun & free_lun, lun
IDL>
IDL> image_data = read_png('out.png')
IDL>
IDL> ; plot image and add text annotation
IDL> p = image(image_data, layout=[5,1,sub_count+1], current=(sub_count gt 0))
IDL> t = text(0,1.1,target=p,"ID="+str(id),color='b',/relative,clip=0)
IDL> endforeach
>> ids = [41092,338375,257378,110568,260067];
>>
>> fig = figure();
>> set(fig,'Position',[0 0 1250 200]);
>>
>> for i=1:numel(ids)
>> url = ['http://www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/' num2str(ids(i))];
>> sub = get_url(url);
>>
>> % it is of course possible this data product does not exist for all requested subhalos
>> if ~isfield(sub.('supplementary_data'),'stellar_mocks'), continue, end
>>
>> % download PNG image, the version which includes all stars in the FoF halo (try replacing 'fof' with 'gz')
>> png_url = sub.('supplementary_data').('stellar_mocks').('image_fof');
>> response = get_url(png_url);
>>
>> % save PNG to temporary file and read it (cannot easily decode PNG format in-memory)
>> f = fopen('out.png','w');
>> fwrite(f,response);
>> fclose(f);
>>
>> image_data = imread('out.png');
>>
>> % plot image and add text annotation
>> subplot(1,numel(ids),i);
>> image(image_data);
>> text(0,-5,['\color{blue} ID=' num2str(ids(i))]);
>> set(gca, 'XTick', []);
>> set(gca, 'YTick', []);
>> end
julia>

Task 11: download the entire Illustris-1 $z=0$ snapshot including only the positions, masses, and metallicities of stars (in the form of 512 HDF5 files).
>>> base_url = "http://www.tng-project.org/api/Illustris-1/"
>>> sim_metadata = get(base_url)
>>> params = {'stars':'Coordinates,Masses,GFM_Metallicity'}
>>>
>>> for i in range(sim_metadata['num_files_snapshot']):
>>> file_url = base_url + "files/snapshot-135." + str(i) + ".hdf5"
>>> saved_filename = get(file_url, params)
>>> print saved_filename
IDL> base_url = "http://www.tng-project.org/api/Illustris-1/"
IDL> sim_metadata = get(base_url)
IDL> params = {stars:'Coordinates,Masses,GFM_Metallicity'}
IDL>
IDL> for i=0,sim_metadata['num_files_snapshot']-1 do begin
IDL> file_url = base_url + "files/snapshot-135." + str(i) + ".hdf5"
IDL> saved_filename = get(file_url, params=params)
IDL> print, saved_filename
IDL> endfor
>> base_url = 'http://www.tng-project.org/api/Illustris-1/';
>> sim_metadata = get_url(base_url);
>> params = struct('stars','Coordinates,Masses,GFM_Metallicity');
>>
>> for i = 1:sim_metadata.('num_files_snapshot')-1
>> file_url = [base_url 'files/snapshot-135.' num2str(i) '.hdf5'];
>> saved_filename = get_url(file_url, params)
>> end
julia>
Note: Extracting only certain particle types or fields from full snapshots
In the above example, since we only need these three fields for stars only, we can reduce the download and storage size from ~1.5TB to ~17GB, in 512 files which are an easy to handle ~35 MB each.Note: Filenames of cutouts and subsets
It is the responsibility of the user to organize the data they download, particularly important when extracting cutouts and subsets. In the above example, the download filenames are completely unmodified -- they have no indication of the fields they contain (this information can be found in a HDF5 Header group). They also do not uniquely identify the files across different simulations. This is also true of halo and subhalo cutouts, whose filenames include only a single ID by default. Therefore, they will collide with different {cutout_query}'s, snapshots, or simulations. The user should implement a directory structure and/or file naming scheme as needed.Task 12: download a particle (gas) cutout around a subhalo and create a synthetic spectral line observation of it, specifically: a HI radio datacube using the MARTINI code. This is an example of tying in TNG data to an external analysis package:
- Jupyter Notebook (note: non-interactive, you can download and run in Lab for interactivity)
Task 13? If you are having trouble figuring out if (or how) a specific task can be accomplished with the API, please request an example!
API Reference
Endpoint Listing and Descriptions
| Endpoint | Description | Return Type |
|---|---|---|
| /api/ | list all simulations currently accessible to the user | json,api (?format=) |
| /api/{sim_name}/ | list metadata (including list of all snapshots+redshifts) for {sim_name} | json,api (?format=) |
| /api/{sim_name}/snapshots/ | list all snapshots which exist for this simulation | json,api (?format=) |
| /api/{sim_name}/snapshots/{num}/ | list metadata for snapshot {num} of simulation {sim_name} | json,api (?format=) |
| /api/{sim_name}/snapshots/z={redshift}/ | redirect to the snapshot which exists closest to {redshift} (with a maximum allowed error of 0.1 in redshift) | json,api (?format=) |
| define [base] = /api/{sim_name}/snapshots/{num} or [base] = /api/{sim_name}/snapshots/z={redshift} (after selection of a particular simulation and snapshot) | ||
| Subfind subhalos | ||
| [base]/subhalos/ | paginated list of all subhalos for this snapshot of this run | json,api (?format=) |
| [base]/subhalos/?{search_query} | execute {search_query} over all subhalos, return those satisfying the search with basic fields and links to /subhalos/{id} | json,api (?format=) |
| [base]/subhalos/plot.pdf?{groupcat_plot_query} | plot the relationship between two or more quantities for (all) subhalos at this snapshot, according to {groupcat_plot_query} | png,jpg.pdf (.ext) |
| [base]/subhalos/{id} | list available data fields and links to all queries possible on subfind subhalo {id} | json,api (?format=) |
| [base]/subhalos/{id}/info.json | extract all group catalog fields for subhalo {id} | json (.ext) |
| [base]/subhalos/{id}/cutout.hdf5 | return snapshot cutout of subhalo {id}, all particle types and fields | HDF5 (.ext) |
| [base]/subhalos/{id}/cutout.hdf5?{cutout_query} | return snapshot cutout of subhalo {id} corresponding to the {cutout_query} | HDF5 (.ext) |
| [base]/subhalos/{id}/vis.png?{vis_query} | return visualization (image/data) of subhalo {id} according to the {vis_query} | png,jpg,pdf,hdf5 (.ext) |
| [base]/subhalos/{id}/skirt/broadband_{survey}.fits | retrieve SKIRT synthetic broadband images FITS file for either SDSS, with {survey}="sdss", or Pan-STARRS with {survey}="pogs". Note: only some subhalos/snapshots, see docs | FITS |
| [base]/subhalos/{id}/skirt/image_{band}_{survey}.png | retrieve SKIRT diagnostic image including morphological details, with {band} = "i" or "g". | PNG |
| FoF halos | ||
| [base]/halos/{halo_id}/ | list what we know about this FoF halo, in particular the 'child_subhalos' | json,api (?format=) |
| [base]/halos/{halo_id}/info.json | extract all group catalog fields for halo {halo_id} | json (.ext) |
| [base]/halos/{halo_id}/cutout.hdf5 | return snapshot cutout of halo {halo_id}, all particle types and fields | HDF5 (.ext) |
| [base]/halos/{halo_id}/cutout.hdf5?{cutout_query} | return snapshot cutout of halo {halo_id} corresponding to the {cutout_query} | HDF5 (.ext) |
| [base]/halos/{halo_id}/vis.png?{vis_query} | return visualization (image/data) of halo {halo_id} according to the {vis_query} | png,jpg,pdf,hdf5 (.ext) |
| merger trees | ||
| [base]/subhalos/{id}/lhalotree/full.hdf5 | retrieve full LHaloTree (flat HDF5 format or hierchical/nested JSON format) | HDF5,json (.ext) |
| [base]/subhalos/{id}/lhalotree/mpb.hdf5 | retrieve only LHaloTree main progenitor branch (towards higher redshift for this subhalo) | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink/full.hdf5 | retrieve full SubLink tree (flat HDF5 format or hierchical/nested JSON format) | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink/mpb.hdf5 | retrieve only SubLink main progenitor branch (towards higher redshift for this subhalo) | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink/mdb.hdf5 | retrieve only SubLink [main] descendant branch(es) (towards lower redshift for this subhalo) (this is the single main branch only if this subhalo lies on the MPB of its z=0 descendant, otherwise it is its full descendant sub-tree containing its z=0 descendant as the first entry) | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink/simple.json | retrieve a simple representation of the SubLink tree, the Main Progenitor Branch (in 'Main') and a list of past mergers (in 'Mergers') only. In both cases a snapshot number and subhalo ID pair is given. | json (.ext) |
| [base]/subhalos/{id}/sublink_gal/full.hdf5 | as above, except for the SubLink_gal tree | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink_gal/mpb.hdf5 | as above, except for the SubLink_gal tree | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink_gal/mdb.hdf5 | as above, except for the SubLink_gal tree | HDF5,json (.ext) |
| [base]/subhalos/{id}/sublink_gal/simple.json | as above, except for the SubLink_gal tree | json (.ext) |
| [base]/subhalos/{id}/sublink/tree.png?{treevis_query} | return visualization of the full SubLink merger tree for subhalo {id} according to the {treevis_query} | png,jpg,pdf (.ext) |
| "on-disk raw" files | ||
| define [base] = /api/{sim_name}/files | ||
| [base]/ | list of each 'files' type available for this simulation (excluding those attached to specific snapshots) | json,api (?format=) |
| [base]/ics.hdf5 | download the initial conditions for this simulation | HDF5 (.ext) |
| [base]/simulation.hdf5 | download the virtual 'simulation.hdf5' container file for this simulation | HDF5 (.ext) |
| [base]/snapshot-{num}/ | list of all the actual file chunks to download snapshot {num} | json,api (?format=) |
| [base]/snapshot-{num}.{chunknum}.hdf5 | download chunk {chunknum} of snapshot {num} | HDF5 (.ext) |
| [base]/snapshot-{num}.{chunknum}.hdf5?{cutout_query} | download only {cutout_query} of chunk {chunknum} of snapshot {num} | HDF5 (.ext) |
| [base]/groupcat-{num}/ | list of all the actual file chunks to download group catalog (fof/subfind) for snapshot {num} | json,api (?format=) |
| [base]/groupcat-{num}/?{subset_query} | download a single field, specified by {subset_query}, from the entire group catalog for snapshot {num} | HDF5 (.ext) |
| [base]/groupcat-{num}.{chunknum}.hdf5 | download chunk {chunknum} of group catalog for snapshot {num} | HDF5 (.ext) |
| [base]/offsets.{num}.hdf5 | download the offsets file for snapshot {num} | HDF5 (.ext) |
| [base]/lhalotree/ | list of all the actual file chunks to download LHaloTree merger tree for this simulation | json,api (?format=) |
| [base]/lhalotree.{chunknum}.hdf5 | download chunk {chunknum} of LHaloTree merger tree for this simulation | HDF5 (.ext) |
| [base]/sublink/ | list of all the actual file chunks to download Sublink merger tree for this simulation | json,api (?format=) |
| [base]/sublink.{chunknum}.hdf5 | download chunk {chunknum} of Sublink merger tree for this simulation | HDF5 (.ext) |
Several API functions accept additional, optional parameters, which are described here.
Search and Cutout requests
{search_query} is an AND combination of restrictions over any of the supported fields, where the
relations supported are 'greater than' (gt), 'greater or equal to' (gte), 'less than' (lt), 'less than or equal to'
(lte), 'equal to'. The first four work by appending e.g. '__gt=val' to the field name (with a double underscore).
For example:
- mass_dm__gt=90.0
- mass__gt=10.0&mass__lte=20.0
- vmax__lt=100.0&len__gas=0&vmaxrad__gt=20.0
{cutout_query} is a concatenated list of particle fields, separated by particle type.
The allowed particle types are 'dm','gas','stars','bhs'. The field names are exactly as in the snapshots
("all" is allowed). Omitting all particle types will return the full cutout: all types, all fields. For example:
- dm=field0,field1,field2&stars=field0,field5,field6&gas=field3
- dm=Coordinates&stars=all
- tracer_tracks=temp (only for the Tracer Tracks supplementary catalog)
{subset_query} is a concatenated list of group catalog fields, separated by object type.
The allowed object types are 'Group' and 'Subhalo'. The field names are exactly as in the group catalogs. Currently, only one single field can be requested at once. For example:
- Group=GroupMass
- Subhalo=SubhaloMassInRadType
- Subhalo=SubhaloSFR
Visualization and Plot requests
{groupcat_plot_query} is a list of "option=value" settings. For example:
- xQuant=mstar2&yQuant=ssfr
- xQuant=mstar_30pkpc&yQuant=delta_sfms&cQuant=fgas
- xQuant=mhalo_500&yQuant=Z_gas&nBins=50&xlim=11.0,15.0&cenSatSelect=sat
| Option Name | Description | Default | Allowed Values |
|---|---|---|---|
| xQuant | x-axis quantity | mstar2_log | any known group catalog quantity (see below) |
| yQuant | y-axis quantity | ssfr | any known group catalog quantity (see below) |
| cQuant | color-axis quantity (by which pixels are colored) | None | if omitted or 'None', then provide a 2d histogram with no third quantity, otherwise any known group catalog quantity (see below) |
| xlim | x-axis limits, automatic if omitted | None | "min,max" pair of floating point numbers e.g. "-3.0,2.0" |
| ylim | y-axis limits, automatic if omitted | None | "min,max" pair of floating point numbers e.g. "-3.0,2.0" |
| clim | color-axis limits, automatic if omitted | None | "min,max" pair of floating point numbers e.g. "-3.0,2.0" |
| cenSatSelect | plot only centrals, satellites, or both? | cen | all, cen, sat |
| cStatistic | statistic to derive color value | median_nan | mean, median, count, sum, median_nan |
| cNaNZeroToMin | set pixels with all NaN/zero values to "minimum" color instead of gray | False | True, False |
| minCount | minimum number of subhalos in a given pixel to include/color in plot | 0 | integer >= zero |
| cRel | if specified, then the color of each pixel represents not the value of cQuant, but instead the relative value of cQuant with respect to the median for subhalos at the same xQuant value (e.g. mass) | None | "min,max" pair of floating point numbers e.g. "0.7,1.3" of the new relative color limits |
| cRelLog | show relative values in log? | False | True, False |
| cFrac | if specified, then the color of each pixel represents not the value of cQuant, but instead the fraction of subhalos in that pixel with a value of cQuant in the given bounds | None | "min,max" pair of floating point numbers e.g. "1.0,inf" (can be unbounded on either side) |
| cFracLog | show fractional values in log? | False | True, False |
| nBins | number of histogram bins | 80 | reasonable integer |
| qRestrict | if specified, a quantity/field to filter on, such that only subhalos which satisfy a value of the field qRestrict within the bounds qRestrictLims are included | None | any known group catalog quantity (see below) |
| qRestrictLims | the required value of the field qRestrict to include subhalos in the plot | None | "min,max" pair of floating point numbers e.g. "-inf,100" (can be unbounded on either side) |
| filterFlag | exclude subhalos which have a bad value of SubhaloFlag? | True | True, False |
| medianLine | draw a median line and [16,84] percentile band? | True | True, False |
| sizeFac | factor to adjust final size/fontsize of plot | 1.0 | floating point number between 0.2 and 4.0 |
| pStyle | plot style, background color choice | white | white, black |
| ctName | colormap/colortable name | gray/viridis | any reasonable colormap name from matplotlib or cmocean. |
The known "group catalog quantities" can be used for (xQuant,yQuant,cQuant,qRestrict). These are:
- None, ssfr, Z_stars, Z_gas, Z_gas_sfr, size_stars, size_gas, fgas1, fgas2, fgas, fdm1, fdm2, fdm, surfdens1_stars, surfdens2_stars, surfdens1_dm, delta_sfms, sfr, sfr1, sfr2, sfr1_surfdens, sfr2_surfdens, virtemp, velmag, spinmag, M_U, M_V, M_B, color_UV, color_VB, vcirc, distance, distance_rvir, zform_mm5, stellarage, massfrac_exsitu, massfrac_exsitu2, massfrac_insitu, massfrac_insitu2, num_mergers, num_mergers_minor, num_mergers_major, num_mergers_major_gyr, mergers_mean_z, mergers_mean_mu, mstar1, mstar2, mstar_30pkpc, mstar_r500, mgas1, mgas2, mhi_30pkpc, mhi2, mgas_r500, fgas_r500, mhalo_200, mhalo_500, mhalo_subfind, mhalo_200_parent, mhalo_vir, halo_numsubs, mstar2_mhalo200_ratio, mstar30pkpc_mhalo200_ratio, rhalo_200, rhalo_500, BH_mass, BH_CumEgy_low, BH_CumEgy_high, BH_CumEgy_ratio, BH_CumEgy_ratioInv, BH_CumMass_low, BH_CumMass_high, BH_CumMass_ratio, BH_Mdot_edd, BH_BolLum, BH_BolLum_basic, BH_EddRatio, BH_dEdt, BH_mode
- Note: this is a very non-exhaustive list of some of the information available on a per-subhalo basis. If you would like to request an addition, which can be derived from the TNG data, please get in touch, and we would be happy to add additional quantities here.
{treevis_query} is a list of "option=value" settings. For example:
- ctName=magma
| Option Name | Description | Default | Allowed Values |
|---|---|---|---|
| ctName | colormap/colortable name | inferno | any reasonable colormap name from matplotlib or cmocean. |
{vis_query} is a list of "option=value" settings. For example:
- partType=gas&partField=coldens
- partType=gas&partField=temp&size=5.0&sizeType=rHalfMassStars
- partType=stars&partField=stellarComp-jwst_f200w-jwst_f115w-jwst_f070w&rotation=face-on&size=50&sizeType=kpc
- partType=dm&partField=velmag
- partType=gas&partField=O VII&method=sphMap
| Option Name | Description | Default | Allowed Values |
|---|---|---|---|
| partType | particle type to visualize | gas | dm, gas, stars |
| partField | field to visualize | coldens_msunkpc2 | any known particle/cell quantity (see below) |
| min | image minimum value | None (automatic) | any floating point value |
| max | image maximum value | None (automatic) | any floating point value |
| rVirFracs | list of "fractions" at which to draw circles denoting the size of the halo/subhalo | 1.0 | a comma-separated list of floating point numbers, e.g. "0.5,1.0,2.0" |
| fracsType | what size measurement should the fractions correspond to? | rVirial | rVirial, rHalfMass, rHalfMassStars, codeUnits, kpc, arcmin |
| method | projection/vis method. Note: "sphMap" includes all FoF particles/cells, while "sphMap_subhalo" includes only subhalo particles/cells. "minIP" and "maxIP" are minimum/maximum intensity projections, respectively. | sphMap_subhalo | sphMap, sphMap_subhalo, sphMap_minIP, sphMap_maxIP, histo |
| nPixels | number of pixels for image | 800,800 | comma-separated integers, e.g. "400,400" or "1920,1080" (maximum: 2000) |
| size | physical image extent (width/height) | 2.5 | any floating point value |
| sizeType | units of 'size' field above | rVirial | rVirial, rHalfMass, rHalfMassStars, codeUnits, kpc, arcmin |
| depthFac | physical image depth (along line of sight), relative to width/height | 1.0 | any floating point value |
| axes | define which axes (of x,y,z) correspond to the image extents, such that the axis omitted becomes the line of sight/projection direction | 0,1 (i.e. "x,y") | "0,1", "0,2", "1,2", "1,0", "2,0", "2,1" |
| axesUnits | unit system for axes labels | code | code, kpc, mpc, deg, arcmin, arcsec |
| labelZ | add redshift label? | True | True, False |
| labelScale | add scalebar label? | True | True, False, physical |
| labelSim | add simulation name label? | True | True, False |
| labelHalo | add object id/mass label? | False | True, False, mstar, mhalo, id, mstar_mhalo |
| plotSubhalos | draw circle markers for the N most massive subhalos of this halo? |
0 | integer >= zero |
| projType | type of projection | ortho | ortho |
| rotation | calculate and apply a rotation based on the galaxy? (leave axes at "0,1") | None | None, face-on, edge-on |
| ctName | colormap/colortable name | (automatic) | any reasonable colormap name from matplotlib or cmocean. |
| plotStyle | plot style, background color choice | open | open, edged, open_black, edged_black |
| rasterPx | number of pixels of output image (if raster), control plot size/fontsize (distinct from nPixels) | 1200 | reasonable integer (maximum: 4000) |
| colorbars | show colorbar? | True | True, False |
| relCoords | show axes distance labels relative to the halo/subhalo? only applies to "open" plot styles | True | True, False |
| title | show plot title? only applies to "open" plot styles | True | True, False |
The known "particle/cell quantities" which can be used for partField are:
- dens, density, mass, masspart, particle_mass, sfr, sfr_msunyrkpc2, coldens, coldens_msunkpc2, coldens_msun_ster, OVI_OVII_ionmassratio, HI, HI_segmented, H2_BR, H2_GK, H2_KMT, HI_BR, HI_GK, HI_KMT, xray, xray_lum, sfr_halpha, halpha, p_sync_ska, temp, temperature, temp_sfcold, ent, entr, entropy, bmag, bmag_uG, bfield_x, bfield_y, bfield_z, dedt, energydiss, shocks_dedt, shocks_energydiss, machnum, shocks_machnum, P_gas, P_B, pressure_ratio, metal, Z, metal_solar, Z_solar, SN_IaII_ratio_Fe, SNIaII_ratio_metals, SN_Ia_AGB_ratio_metals, vmag, velmag, vel_los, vel_los_sfrwt, vel_x, vel_y, vel_z, velsigma_los, velsigma_los_sfrwt, vrad, halo_vrad, radvel, halo_radvel, vrad_vvir, specangmom_mag, specj_magstar_age, stellar_age, stellarComp-jwst_f200w-jwst_f115w-jwst_f070w, potential, id, metals_H, metals_He, metals_C, metals_N, metals_O, metals_Ne, metals_Mg, metals_Si, metals_Fe, {ionName}, {ionName} fracmass, {ionName} mass, sb_{emLine}, stellarBand-{bandName}, stellarBandObsFrame-{bandName}
- {ionName} options: Al I, Al II, Al III, Al IV, Al V, Al VI, Al VII, Al VIII, Al IX, Al X, Ar I, Ar II, Ar III, Ar IV, Ar V, Ar VI, Ar VII, Ar VIII, Ar IX, Ar X, Be I, Be II, Be III, Be IV, Be V, B I, B II, B III, B IV, B V, B VI, Ca I, Ca II, Ca III, Ca IV, Ca V, Ca VI, Ca VII, Ca VIII, Ca IX, Ca X, C I, C II, C III, C IV, C V, C VI, C VII, Cl I, Cl II, Cl III, Cl IV, Cl V, Cl VI, Cl VII, Cl VIII, Cl IX, Cl X, Cr I, Cr II, Cr III, Cr IV, Cr V, Cr VI, Cr VII, Cr VIII, Cr IX, Cr X, Co I, Co II, Co III, Co IV, Co V, Co VI, Co VII, Co VIII, Co IX, Co X, Cu I, Cu II, Cu III, Cu IV, Cu V, Cu VI, Cu VII, Cu VIII, Cu IX, Cu X, F I, F II, F III, F IV, F V, F VI, F VII, F VIII, F IX, F X, He I, He II, He III, H I, H II, H III, Fe I, Fe II, Fe III, Fe IV, Fe V, Fe VI, Fe VII, Fe VIII, Fe IX, Fe X, Li I, Li II, Li III, Li IV, Mg I, Mg II, Mg III, Mg IV, Mg V, Mg VI, Mg VII, Mg VIII, Mg IX, Mg X, Mn I, Mn II, Mn III, Mn IV, Mn V, Mn VI, Mn VII, Mn VIII, Mn IX, Mn X, Ne I, Ne II, Ne III, Ne IV, Ne V, Ne VI, Ne VII, Ne VIII, Ne IX, Ne X, Ni I, Ni II, Ni III, Ni IV, Ni V, Ni VI, Ni VII, Ni VIII, Ni IX, Ni X, N I, N II, N III, N IV, N V, N VI, N VII, N VIII, O I, O II, O III, O IV, O V, O VI, O VII, O VIII, O IX, P I, P II, P III, P IV, P V, P VI, P VII, P VIII, P IX, P X, K I, K II, K III, K IV, K V, K VI, K VII, K VIII, K IX, K X, Sc I, Sc II, Sc III, Sc IV, Sc V, Sc VI, Sc VII, Sc VIII, Sc IX, Sc X, Si I, Si II, Si III, Si IV, Si V, Si VI, Si VII, Si VIII, Si IX, Si X, Na I, Na II, Na III, Na IV, Na V, Na VI, Na VII, Na VIII, Na IX, Na X, S I, S II, S III, S IV, S V, S VI, S VII, S VIII, S IX, S X, Ti I, Ti II, Ti III, Ti IV, Ti V, Ti VI, Ti VII, Ti VIII, Ti IX, Ti X, V I, V II, V III, V IV, V V, V VI, V VII, V VIII, V IX, V X, Zn I, Zn II, Zn III, Zn IV, Zn V, Zn VI, Zn VII, Zn VIII, Zn IX, Zn X
- {emLine} options: H--1-911.753A, H--1-3645.98A, H--1-1215.67A, H--1-1025.72A, H--1-972.537A, H--1-6562.81A, H--1-4861.33A, H--1-4340.46A, H--1-4101.73A, C--6-33.7372A, N--7-24.7807A, O--8-18.9709A, O--8-16.0067A, O--8-15.1767A, O--8-102.443A, O--8-75.8835A, Ne10-12.1375A, Na11-10.0250A, Mg12-8.42141A, Si14-6.18452A, S-16-4.73132A, Fe26-1.78177A, C--5-40.2678A, C--5-41.4721A, N--6-29.5343A, O--7-21.8070A, O--7-21.8044A, O--7-21.6020A, O--7-22.1012A, Ne-9-13.6987A, Ne-9-13.5529A, Ne-9-13.5500A, Ne-9-13.4471A, Mg11-9.31434A, Mg11-9.23121A, Mg11-9.22816A, Mg11-9.16875A, Si13-6.74039A, Si13-6.68828A, Si13-6.68508A, Si13-6.64803A, S-15-5.10150A, S-15-5.06649A, S-15-5.06314A, S-15-5.03873A, Fe25-1.86819A, Fe25-1.85951A, Fe25-1.85541A, Fe25-1.85040A, C--3-1908.73A, C--3-1906.68A, C--3-977.020A, C--4-1550.78A, C--4-1548.19A, O--6-1037.62A, O--6-1031.91A, O--6-183.937A, O--6-184.117A, S--4-1404.81A, S--4-1423.84A, S--4-1398.04A, O--2-3728.81A, O--2-3726.03A, O--3-4931.23A, O--3-4958.91A, O--3-5006.84A, N--2-6527.23A, N--2-6548.05A, N--2-6583.45A, S--2-6730.82A, S--2-6716.44A, S--2-4076.35A, S--2-4068.60A, O--6-5291.00A, O--6-2082.00A, O--6-3434.00A, O--6-2070.00A, O--6-1125.00A, Blnd-2798.00A, Blnd-1035.00A, Lyman-alpha, Lyman-beta, MgII, H-alpha, H-beta, [OII]3729, OVII, OVIII
- {bandName} options: wfpc2_f439w, isaac_ks, iras_12, jwst_f356w, scuba_450wb, wfc3_uvis_f218w, pacs_100, buser_b, i2300, wfc3_ir_f140w, wfc_acs_f814w, wfc3_uvis_f814w, nicmos_f160w, wfcam_k, steidel_un, cousins_r, iras_25, wfcam_z, wfcam_y, wfc_acs_f625w, wfc3_ir_f098m, cousins_i, newfirm_j1, newfirm_j3, i1500, newfirm_j2, wise_w1, ps1_i, wise_w3, wise_w2, wise_w4, bessell_l, bessell_m, ps1_r, ps1_y, ps1_z, jwst_f277w, sdss_u, des_r, des_y, des_z, bessell_lp, wfc3_uvis_f438w, 2mass_ks, steidel_rs, des_g, des_i, uvot_m2, wfc3_ir_f110w, stromgren_u, stromgren_v, wfc_acs_f850lp, wfc3_uvis_f390w, stromgren_y, stromgren_b, wfpc2_f850lp, wfc3_uvis_f336w, spire_500, wfpc2_f555w, fors_v, uvot_w2, fors_r, wfc3_uvis_f606w, iras_100, newfirm_h2, newfirm_h1, wfc_acs_f475w, wfpc2_f814w, jwst_f070w, mips_24, wfc3_uvis_f775w, suprimecam_r, suprimecam_v, suprimecam_i, wfc_acs_f435w, suprimecam_b, mips_70, 2mass_h, 2mass_j, sdss_g, wfpc2_f300w, sdss_i, wfc3_uvis_f475w, sdss_r, wfc3_uvis_f850lp, newfirm_k, sdss_z, wfc3_ir_f160w, wfc_acs_f555w, wfc3_ir_f125w, i2800, vista_y, megacam_u, megacam_r, suprimecam_z, wfpc2_f450w, wfc3_uvis_f225w, wfc_acs_f606w, galex_nuv, vista_h, vista_k, vista_j, wfpc2_f336w, megacam_i, b, wfc3_ir_f105w, galex_fuv, mips_160, v, uvot_w1, jwst_f444w, wfc3_uvis_f555w, wfc3_uvis_f275w, wfcam_j, spire_350, wfcam_h, jwst_f200w, pacs_160, jwst_f150w, wfpc2_f255w, wfc_acs_f775w, jwst_f115w, spire_250, scuba_850wb, wfpc2_f606w, ps1_g, iras_60, nicmos_f110w, pacs_70, megacam_z, irac_4, irac_2, irac_3, irac_1, megacam_g, cfht_i, suprimecam_g, cfht_b, u, steidel_i, steidel_g, jwst_f090w, cfht_r, lsst_u, lsst_g, lsst_r, lsst_i, lsst_z, lsst_z, lsst_y, euclid_vis, euclid_y, euclid_j, euclid_h, roman_f062, roman_f087, roman_f106, roman_f129, roman_f158, roman_f184
[base]/subhalos/{id} fields
The endpoint for an individual subhalo returns many numeric fields, as well as links to additional data, when available:
- All Subfind group catalog fields are given, flattened into separate values if a vector,
and with modified names. In particular, the 'Subhalo' prefix is removed. For values by
type, the 'Type' postfix is removed from the end of the name and '_gas', '_dm', '_stars', and
'_bhs' added. For 3-vectors, '_x', '_y', and '_z' are added. For example,
SubhaloPosbecomespos_x,pos_y,pos_z. - Main branch merger tree links from SubLink are given in terms of their Subfind IDs and snapshots.
For first progenitors, the fields are
prog_sfidandprog_snap. For descendants, the fields aredesc_sfidanddesc_snap. If no such relation exists, both values equal -1. - The
supplementary_datafield contains any pre-computed data products which are available for this subhalo (may be empty).
Response codes:
The Illustris API will return a subset of standard HTTP response codes, in particular:
- 200 - Success. response is being returned as requested.
- 302 - Redirection. client must submit a second, otherwise identical request to the new URL specified in the Location header field. (All browsers, libraries and commands will do this automatically and transparently). For example, all raw file download requests will receive a redirect type response.
- 400 - Bad Request. The server will not process the request, because it appears to contain an error. For example, we return code 400 if you make a cutout request but include a particle type or field name which is not known (e.g. typo).
- 401 - Unauthorized. Standard response requesting authentication if it was not provided. You will receive this response if you do not include your API Key with a request.
- 403 - Permission Denied. Standard response for insufficient privileges, when you have authenticated successfully.
- 404 - Not Found. Either there is no API endpoint at the URL which you requested, or you have requested a data resource which does not exist. For example, requesting a 'stellar mocks' broadband file for a subhalo for which it is not currently available.
- 500 - Server Error. Our fault, this is not good! Please let us know if this happens.
Specifying request formats
Each API eachpoint can return a response in one or more data types. When multiple options exist, a specify return format can be requested through one of the following methods.
-
(?format=)indicates that the return type is chosen by supplying such a querystring, appended to the URL. -
(.ext)indicates that the return type is chosen by supplying the desired file extension in the URL.
Sending authentication
All API requests require authentication, and therefore also user registration. Each request must provide, along with the details of the request itself, the unique "API Key" of the user making the request. You can send your API Key in two ways:
- in the querystring, by appending it to the URL (the following are just examples,
replace with your actual key):
?api_key=INSERT_API_KEY_HERE. - in HTTP header. This is particularly useful for wget commands or
within scripts:
api-key: INSERT_API_KEY_HERE.
Note: If you are logged in to the website, then requests from your browser are automatically authenticated. Navigating the Browsable API works in this way.
Full-path examples
To be explicit by way of example, the following are absolute URLs for the Illustris API covering some of its functionality, where the type of the request and the expected response should be clear from the preceding documentation.
- www.tng-project.org/api/Illustris-2/
- www.tng-project.org/api/Illustris-2/snapshots/68/
- www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/73664/
- www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/73664/stellar_mocks/broadband.fits
- www.tng-project.org/api/Illustris-1/snapshots/135/subhalos/73664/stellar_mocks/sed.txt
- www.tng-project.org/api/Illustris-1/snapshots/80/halos/523312/cutout.hdf5?dm=Coordinates,Velocities&gas=Coordinates,Masses
- www.tng-project.org/api/Illustris-3/snapshots/135/subhalos?mass__gt=10.0&mass__lt=20.0
- www.tng-project.org/api/Illustris-2/snapshots/68/subhalos/50000/sublink/full.hdf5
- www.tng-project.org/api/Illustris-2/snapshots/68/subhalos/50000/sublink/mpb.json
- www.tng-project.org/api/TNG100-2/snapshots/99/subhalos/50000/sublink/mpb.json
- www.tng-project.org/api/TNG300-1/snapshots/99/subhalos?mass__gt=10.0&mass__lt=11.0
- www.tng-project.org/api/Illustris-1/files/groupcat-135.5.hdf5
- www.tng-project.org/api/Illustris-1/files/snapshot-135.200.hdf5
- www.tng-project.org/api/Illustris-2/files/snapshot-135.10.hdf5?dm=all
- www.tng-project.org/api/Illustris-3/files/sublink.2.hdf5
- www.tng-project.org/api/Illustris-1/files/groupcat-135/?Subhalo=SubhaloMass
- www.tng-project.org/api/TNG100-1/snapshots/50/subhalos/plot.png?xQuant=mstar2_log&yQuant=ssfr
- www.tng-project.org/api/TNG300-1/snapshots/50/subhalos/plot.png?xQuant=mstar2_log&yQuant=delta_sfms&cQuant=Z_gas
- www.tng-project.org/api/TNG100-1/snapshots/99/halos/320/vis.png
- www.tng-project.org/api/TNG100-2/snapshots/67/halos/0/vis.png?partType=gas&partField=temp