Using the Tracers (finding parents at earlier snapshots)
Josue Lopez
22 Jan '24
Hello,
I am currently working with galaxy data in the TNG-50 simulation and would like to trace the origin of the gas. Specifically, I am interested in determining whether it comes from the intergalactic medium or from another galaxy, and if so, which one. While I have explored the tracers, I am having difficulty loading the necessary data. It appears that the files are empty, but I am not sure if I am doing it correctly. Any help would be greatly appreciated.
Dylan Nelson
23 Jan '24
Are you loading them yourself from the snapshot files? Perhaps you can paste a code snippet.
Josue Lopez
24 Jan '24
Thank you for your response. Firstly, I printed the number of particles for a subhalo in TNG50. The output showed that there was no data for Particle Type 3, which I understand corresponds to the tracers.
This is my code:
I also tried using the dataset mentioned in index a) of the Data Specification. From the information provided, I understand that to trace the gas to a galaxy, we need to use the file 'tr_all_groups_99_subhalo_id'. I went to the path 'TNG/TNG50-1/postprocessing/tracer_tracks' to find the hdf5 file, but I only found two files: 'tr_all_groups_99_meta.hdf5' and 'tr_all_groups_99_parent_indextype.hdf5'. I do not understand how to trace the origin of the gas using these two files. Thanks in advance for your help.
Here is the other part of my code:
basePath = '../sims.TNG/TNG50-1/postprocessing/tracer_tracks'
file = basePath+"/tr_all_groups_99_meta.hdf5"
data = h5py.File(file,"r")
print("####Data")
for f in data.keys():
print(f)
print("\n")
print(np.array(data["Halo/TracerLength/gas"]))
Dylan Nelson
24 Jan '24
Hi Josue,
I would suggest to read the data release papers, which describe the tracers a bit more. They are not so easy to work with. They are not ordered by (sub)halo, so (sub)halos do not "contain" any tracers. To load and use them, you have to load all PartType3 from the snapshot using ill.snapshot.loadSubset(), for example.
The supplementary data catalog "(a) Tracer Tracks" is a good alternative, it makes them easier to use. The documentation gives some pseudo-code advice on how to use this dataset.
Guan-Fu Liu
6 May '25
Hi!
I am also trying to deal with the '/home/tnguser/sims.TNG/TNG50-1/postprocessing/tracer_tracks/tr_all_groups_99_meta.hdf5' and I find some difficulties. Before describing my questions in detail, I would like to paste some code snippet
basePath = '/home/tnguser/sims.TNG/TNG50-1/postprocessing/tracer_tracks'
file = '%s/tr_all_groups_99_meta.hdf5' % basePath
f = h5py.File(file)
print('Keys of f:', f.keys())
print("Keys of f['Subhalo']:", f['Subhalo'].keys())
print("Keys of f['Subhalo']['TracerOffset']:", f['Subhalo']['TracerOffset'].keys())
print("Keys of f['Subhalo']['TracerLength']:", f['Subhalo']['TracerLength'].keys())
start = f['Subhalo']['TracerOffset']['gas'][6]
length = f['Subhalo']['TracerLength']['gas'][6]
parentIDs = f['ParentIDs'][start:start+length]
tracerIDs = f['TracerIDs'][start:start+length]
print('shape of parentIDs:', parentIDs.shape)
print('shape of tracerIDs:', tracerIDs.shape)
and the printed output is
Keys of f: <KeysViewHDF5 ['Halo', 'ParentIDs', 'Subhalo', 'TracerIDs']>
Keys of f['Subhalo']: <KeysViewHDF5 ['TracerLength', 'TracerOffset']>
Keys of f['Subhalo']['TracerOffset']: <KeysViewHDF5 ['bhs', 'gas', 'stars']>
Keys of f['Subhalo']['TracerLength']: <KeysViewHDF5 ['bhs', 'gas', 'stars']>
shape of parentIDs: (81,)
It seems good and I obtain the parentIDs and tracerIDs of tracer particles whose parent particles are gas in subhalo 6 at snap=99, my questions are
(1) Do I obtain the parentIDs and tracerIDs of tracer particles whose parent particles are gas in subhalo 6 at snap=99 correctly?
(2) If I load the the parentIDs and tracerIDs of tracer particles whose parent particles are gas in subhalo 6 at snap=99 correctly, how can I get their properties like postions, masses, and etc.? I am planning to load all the gas particles of snap=99 via gas=il.snapshot.loadSubset(basePath, 99, 'gas', fields=['particleIDs','Masses'], subset=None) and only select the gas particles whose particleIDs match the tracer particles in subhalo 6 at snap=9. Such a method seems to be slow, is there any faster way?
(3) If method is step (2) are correct and the fastest way, how can I get the properties of tracer particles at snap=98 or earlier? There is only tr_all_groups_99_meta.hdf5, no files like tr_all_groups_98_meta.hdf5 nor tr_all_groups_97_meta.hdf5, etc.
Many thanks!
Dylan Nelson
6 May '25
(1) That's correct.
(2) That's also correct, in general. To select the gas with matching IDs, an algorithm based on sorting is a good approach, you can use something like np.intersect1d.
However, using the "tracer_tracks", you don't actually need to do this (see below).
(3) The other file, parent_indextype, solves this problem, as well as the problem above. This is because it gives you immediately the index (not the ID) of the parent, at all previous snapshots.
Given a list of snapshot indices, you can either load corresponding fields by calling il.snapshot.loadSubset() and passing subset (see this thread for the idea). Or, you can simply use the simulation.hdf5 file like
with h5py.File(basePath + 'simulation.hdf5','r') as f:
pos = f['/Snapshots/98/PartType0/Coordinates'][indices]
Guan-Fu Liu
6 May '25
Thanks!
I try the other file, and the following are my codes and outputs:
basePath = '/home/tnguser/sims.TNG/TNG50-1/postprocessing/tracer_tracks'
file1 = '%s/tr_all_groups_99_parent_indextype.hdf5' % basePath
f1 = h5py.File(file1)
print('Keys of f1:', f1.keys())
print("Shape of f1['parent_indextype']:", f1['parent_indextype'].shape)
print("Shape of f1['redshifts']:", f1['redshifts'].shape)
print("Shape of f1['snaps']:", f1['snaps'].shape)
print("f1['parent_indextype'][:10,0]:",f1['parent_indextype'][:10,0])
indices = f1['parent_indextype'][:10,0]
print("f['snap'][0]:", f1['snaps'][0])
Keys of f1: <KeysViewHDF5 ['parent_indextype', 'redshifts', 'snaps']>
Shape of f1['parent_indextype']: (3292847014, 100)
Shape of f1['redshifts']: (100,)
Shape of f1['snaps']: (100,)
f1['parent_indextype'][:10,0]: [0 2 2 3 3 4 4 4 4 5]
f['snap'][0]: 99
It confuses me that why there are some repeating numbers in f1['parent_indextype'][:10,0]? I guess that there are multiple progenitor particles that contribute to the tracer particles at snapNum=99.
Dylan Nelson
6 May '25
A parent can always have multiple tracers.
In this case, gas cell indices 2 and 3 both have two tracer children.
Guan-Fu Liu
7 May '25
To check something more, I think f1['parent_indextype'][:10,0]: [0 2 2 3 3 4 4 4 4 5] should be interpreted as follows,
Tracer with a tracerID of 0 at snapNum=99, corresponds to the particle with a particleID of 0;
Tracer with a tracerID of 1 and 2 at snapNum=99, corresponds to the particle with a particleID of 2;
Tracer with a tracerID of 3 and 4 at snapNum=99, corresponds to the particle with a particleID of 3;
Tracer with a tracerID of 5, 6, 7, 8 at snapNum=99, corresponds to the particle with a particleID of 4;
Tracer with a tracerID of 9 at snapNum=99, corresponds to the particle with a particleID of 5.
If so here comes the problem, how can I know the part type of the particles with the particleIDs that was mentioned above.
Dylan Nelson
7 May '25
(1) Please see the definition of "parent_indextype", this is a number that represents both the type and index.
(2) Don't mix up TracerID and index, your first numbers "0", "1 and 2", "3 and 4" and so on, are the indices of the tracers, not their IDs.
(3) "corresponds to the particle" should be "corresponds to the gas cell". This is because all the parents of these tracers have type 0. This is because the "parent_indextype" values are less than 1e11 (see definition).
(4) "with a particle ID of..." should be "with the index of..." (particles and gas cells have IDs, and indices, and these numbers give the indices).
Guan-Fu Liu
18 May '25
Hi! I found that it may be more easier for me to load the whole tracer particles at some given snapshots and I still met with some questions, which are described as follows:
Here is my code
tracer = il.snapshot.loadSubset(basePath, 99, 'tracer')
print('There are %d tracer at z=0'%tracer['count'])
print(tracer['TracerID'].min(), tracer['TracerID'].max(), len(np.unique(tracer['TracerID'])))
tracer = il.snapshot.loadSubset(basePath, 33, 'tracer')
print('There are %d tracer at z=2.0'%tracer['count'])
print(tracer['TracerID'].min(), tracer['TracerID'].max(), len(np.unique(tracer['TracerID'])))
the output is
There are 1487761408 tracer at z=0
100000000010 110077695995 1487761408
There are 1487761408 tracer at z=2.0
100000000002 110077695997 1487761408
My questions are:
(1) Will some tracers disappear across different snapshots? From the output, the tracer particle with a TracerID of 100000000002 does not exist at the snapshot z=0.0.
(2) If the answer of (1) is yes, does it because some physical processes like SMBH accretion?
(3) How does the simulation generate new tracers?
Many thanks!
Dylan Nelson
19 May '25
(1) No, by definition tracers never appear or disappear.
In TNG50-1, at z=0, the minimum PartType3/TracerID is also 100000000002, so perhaps there is an issue in your loading code above. Could be subtle, i.e. related to memory? Since these are large arrays.
(2) It doesn't. Tracers are generated in the ICs (at t=0) only.
Guan-Fu Liu
19 May '25
Hi! I re-run some similar codes in the TNG officical JupterLab Workspace, and the complete code is as follows,
# Basic packages about scientific computation
import numpy as np
# Packages fomr IllustrisTNG official
import illustris_python as il
# h5 file handling
import h5py
import copy
import gc
basePath = '/home/tnguser/sims.TNG/TNG50-1/output'
PartTypeIndex = {"gas":0, "dm":1, "Unused":2, "tracers":3, "stars":4, "BHs":5} # Mass Types
file = il.snapshot.snapPath(basePath, 99)
with h5py.File(file, 'r'):
f = h5py.File(file, 'r')
header = dict(f['Header'].attrs.items())
nTracer = header['NumPart_Total'][PartTypeIndex['tracers']]
nChunks = np.array([0, nTracer // 4*1, nTracer // 4*2, nTracer // 4*3, nTracer], dtype=np.uint64)
subset1 = il.snapshot.getSnapOffsets(basePath, 99, 0, "Subhalo")
miniIDs = [ ]
for i in range(len(nChunks)):
if i==(len(nChunks)-1):
continue
subset = copy.deepcopy(subset1)
subset['lenType'][PartTypeIndex['tracers']] = nChunks[i+1] - nChunks[i]
subset['offsetType'][PartTypeIndex['tracers']] = nChunks[i]
tracer = il.snapshot.loadSubset(basePath, 99, 'tracer', subset=subset)
print('Chunk %d, the minimum TracerID is %d'%(i, tracer['TracerID'].min()))
miniIDs.append(tracer['TracerID'].min())
del subset
del tracer
print('%d chunks were done' % (i+1))
gc.collect()
print('At z=0, the minimum TracerIDs at 4 chunks are', miniIDs)
the output of snapshot at z=0 is
Chunk 0, the minimum TracerID is 100000000187
1 chunks were done
Chunk 1, the minimum TracerID is 100000000068
2 chunks were done
Chunk 2, the minimum TracerID is 100000000010
3 chunks were done
Chunk 3, the minimum TracerID is 100000000075
4 chunks were done
At z=0, the minimum TracerIDs at 4 chunks are [100000000187, 100000000068, 100000000010, 100000000075]
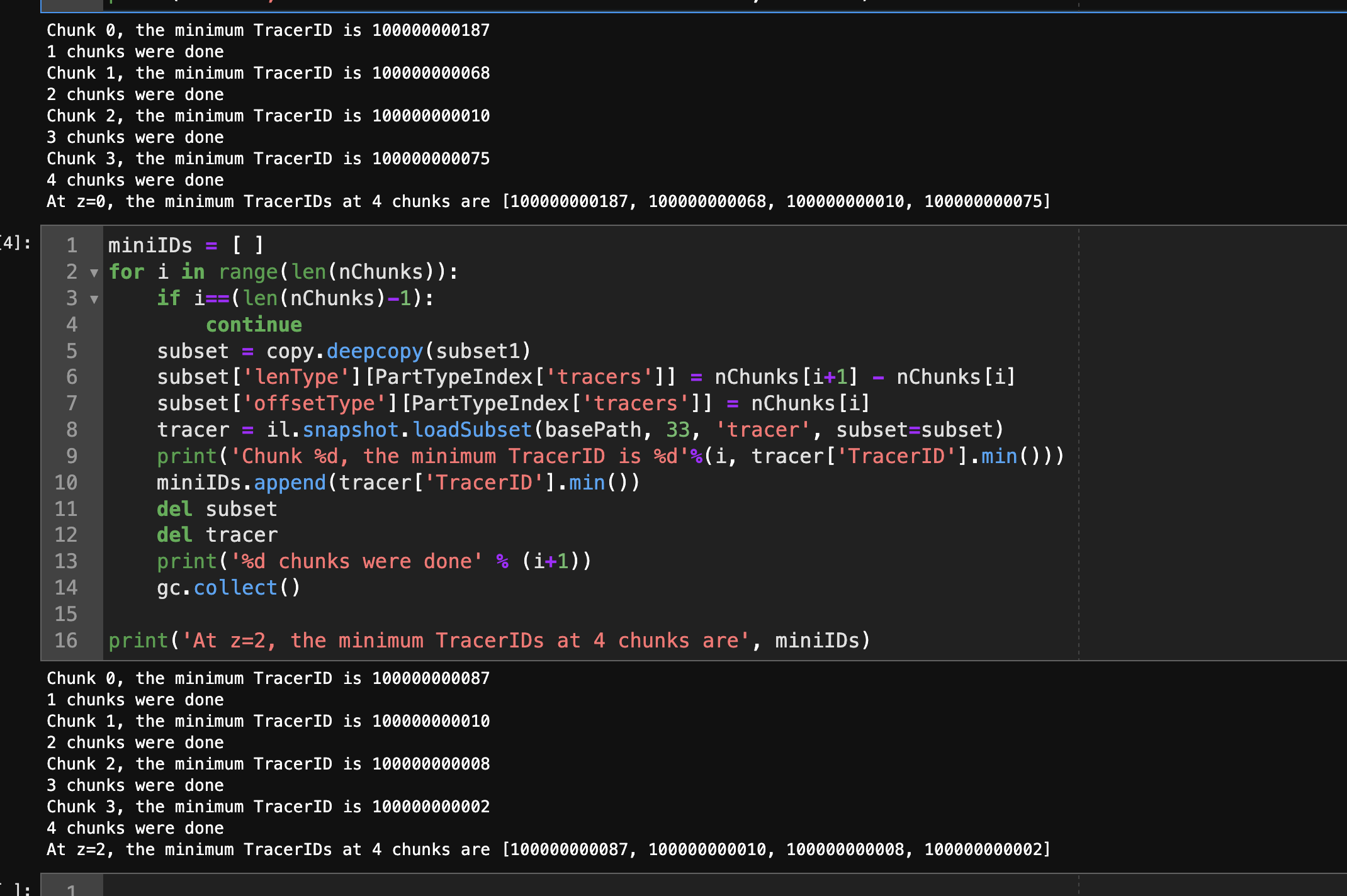
I also run the similar one but for z=2
miniIDs = [ ]
for i in range(len(nChunks)):
if i==(len(nChunks)-1):
continue
subset = copy.deepcopy(subset1)
subset['lenType'][PartTypeIndex['tracers']] = nChunks[i+1] - nChunks[i]
subset['offsetType'][PartTypeIndex['tracers']] = nChunks[i]
tracer = il.snapshot.loadSubset(basePath, 33, 'tracer', subset=subset)
print('Chunk %d, the minimum TracerID is %d'%(i, tracer['TracerID'].min()))
miniIDs.append(tracer['TracerID'].min())
del subset
del tracer
print('%d chunks were done' % (i+1))
gc.collect()
print('At z=2, the minimum TracerIDs at 4 chunks are', miniIDs)
the output of snapshot at z=2.0 is
Chunk 0, the minimum TracerID is 100000000087
1 chunks were done
Chunk 1, the minimum TracerID is 100000000010
2 chunks were done
Chunk 2, the minimum TracerID is 100000000008
3 chunks were done
Chunk 3, the minimum TracerID is 100000000002
4 chunks were done
At z=2, the minimum TracerIDs at 4 chunks are [100000000087, 100000000010, 100000000008, 100000000002]
It's a bit complex, I would suggest a simpler test:
cd sims.TNG/TNG50-1/output/snapdir_099/
minmax = [np.inf, 0.0]
for i in range(680):
with h5py.File('snap_099.%d.hdf5' % i,'r') as f:
ids = f['PartType3']['TracerID'][()]
minmax[0] = np.min([minmax[0],np.min(ids)])
minmax[1] = np.max([minmax[1],np.max(ids)])
print(i)
print(f'{minmax = }')
Note: the min/max in any particular chunk is not important, only the global (across all chunks) is.
Guan-Fu Liu
19 May '25
Got it! But the global minimum TracerID at z=2.0 and z=0 is still different. In the previous code, I run it in my local machine via load the tracers of the whole snapshot. In the latter code, I run it in TNG JupterLab where I only have a memory no more than 10GB and must split them in chunks. Both of them give a different global minmum TracerID at z=2.0 and z=0.
Guan-Fu Liu
19 May '25
Hi! I think I locate the problem.
In the last two kinds of codes mentioned above, both of them use il.snapshot.loadSubset. In the last code you gave, it uses h5py to load the snapshot files. I also try to load the snapshot file via h5py directly and found the minimum and maximum values of TracerID across snapshots are the same.
In short, do not use il.snapshot.loadSubset to load tracer particles and you had better to load it via h5py.
Guan-Fu Liu
19 May '25
Hi! I guess that I figured the reason: il.snapshot.loadSubset will use the number, header['NumPart_Total'][PartTypeIndex['tracers']], which has a value of 1487761408. However, there is 10077696000 tracer particles. Such a larger number can also be found in the url TNG50-1 Data ana be obtained by adding up the number header['NumPart_ThisFile'][PartTypeIndex['tracers']] of the total 680 files (for TNG50-1).
It still confuses me why there are these two numbers.
Dylan Nelson
19 May '25
Due to the limits on 32 bit numbers, slightly more work is needed to obtain total particle counts from the headers, as implemented in the getNumPart() function.
Got it! After some tests, I found that there may be a bug here. Let me explain it step by step:
The original code of getNumPart is
def getNumPart(header):
""" Calculate number of particles of all types given a snapshot header. """
nTypes = 6
nPart = np.zeros(nTypes, dtype=np.int64)
for j in range(nTypes):
nPart[j] = header['NumPart_Total'][j] | (header['NumPart_Total_HighWord'][j] << 32)
return nPart
The header['NumPart_Total_HighWord'] of TNG 50-1 is array([2, 2, 0, 2, 0, 0], dtype=uint32)
The header['NumPart_Total'] of TNG50-1 is array([ 802737432, 1487761408, 0, 1487761408, 341411850, 3028], dtype=uint32)
For tracer particles, j=3 in getNumPart, and the total number of tracer particles is header['NumPart_Total_HighWord'][3] *2**32 + header['NumPart_Total'][3], which is 2^32 + 1487761408.
However, in getNumPart, the data type of header['NumPart_Total_HighWord'][3] is np.uint32, so header['NumPart_Total_HighWord'][3] *2**32(header['NumPart_Total_HighWord'][3] <<32 exceeds the maximum limit of np.uint32, then header['NumPart_Total_HighWord'][3] <<32 is 0, the total number returned by getNumPart is wrong.
To solve it, we may need change the line in getNumPart
I found that the code in getNumPart function solved such a bug 10 months ago. I was provided the account of TNG JupyterLab about 3 years ago, so the package is of the older version.
Dylan Nelson
20 May '25
Hi yes I see, that seems correct.
Unfortunately you're right - Lab accounts are given some starting files when they are created, and these are not automatically updated. This situation could be improved.
You can delete your illustris_python and get the newest version from github.
Hello,
I am currently working with galaxy data in the TNG-50 simulation and would like to trace the origin of the gas. Specifically, I am interested in determining whether it comes from the intergalactic medium or from another galaxy, and if so, which one. While I have explored the tracers, I am having difficulty loading the necessary data. It appears that the files are empty, but I am not sure if I am doing it correctly. Any help would be greatly appreciated.
Are you loading them yourself from the snapshot files? Perhaps you can paste a code snippet.
Thank you for your response. Firstly, I printed the number of particles for a subhalo in TNG50. The output showed that there was no data for Particle Type 3, which I understand corresponds to the tracers.
This is my code:
I also tried using the dataset mentioned in index a) of the Data Specification. From the information provided, I understand that to trace the gas to a galaxy, we need to use the file 'tr_all_groups_99_subhalo_id'. I went to the path 'TNG/TNG50-1/postprocessing/tracer_tracks' to find the hdf5 file, but I only found two files: 'tr_all_groups_99_meta.hdf5' and 'tr_all_groups_99_parent_indextype.hdf5'. I do not understand how to trace the origin of the gas using these two files. Thanks in advance for your help.
Here is the other part of my code:
Hi Josue,
I would suggest to read the data release papers, which describe the tracers a bit more. They are not so easy to work with. They are not ordered by (sub)halo, so (sub)halos do not "contain" any tracers. To load and use them, you have to load all PartType3 from the snapshot using
ill.snapshot.loadSubset(), for example.The supplementary data catalog "(a) Tracer Tracks" is a good alternative, it makes them easier to use. The documentation gives some pseudo-code advice on how to use this dataset.
Hi!
I am also trying to deal with the
'/home/tnguser/sims.TNG/TNG50-1/postprocessing/tracer_tracks/tr_all_groups_99_meta.hdf5'and I find some difficulties. Before describing my questions in detail, I would like to paste some code snippetand the printed output is
It seems good and I obtain the parentIDs and tracerIDs of tracer particles whose parent particles are gas in subhalo 6 at snap=99, my questions are
gas=il.snapshot.loadSubset(basePath, 99, 'gas', fields=['particleIDs','Masses'], subset=None)and only select the gas particles whose particleIDs match the tracer particles in subhalo 6 at snap=9. Such a method seems to be slow, is there any faster way?tr_all_groups_99_meta.hdf5, no files liketr_all_groups_98_meta.hdf5nortr_all_groups_97_meta.hdf5, etc.Many thanks!
(1) That's correct.
(2) That's also correct, in general. To select the gas with matching IDs, an algorithm based on sorting is a good approach, you can use something like np.intersect1d.
However, using the "tracer_tracks", you don't actually need to do this (see below).
(3) The other file,
parent_indextype, solves this problem, as well as the problem above. This is because it gives you immediately the index (not the ID) of the parent, at all previous snapshots.Given a list of snapshot indices, you can either load corresponding fields by calling
il.snapshot.loadSubset()and passingsubset(see this thread for the idea). Or, you can simply use thesimulation.hdf5file likeThanks!
I try the other file, and the following are my codes and outputs:
It confuses me that why there are some repeating numbers in f1['parent_indextype'][:10,0]? I guess that there are multiple progenitor particles that contribute to the tracer particles at snapNum=99.
A parent can always have multiple tracers.
In this case, gas cell indices 2 and 3 both have two tracer children.
To check something more, I think
f1['parent_indextype'][:10,0]: [0 2 2 3 3 4 4 4 4 5]should be interpreted as follows,If so here comes the problem, how can I know the part type of the particles with the particleIDs that was mentioned above.
(1) Please see the definition of "parent_indextype", this is a number that represents both the type and index.
(2) Don't mix up TracerID and index, your first numbers "0", "1 and 2", "3 and 4" and so on, are the indices of the tracers, not their IDs.
(3) "corresponds to the particle" should be "corresponds to the gas cell". This is because all the parents of these tracers have type 0. This is because the "parent_indextype" values are less than 1e11 (see definition).
(4) "with a particle ID of..." should be "with the index of..." (particles and gas cells have IDs, and indices, and these numbers give the indices).
Hi! I found that it may be more easier for me to load the whole tracer particles at some given snapshots and I still met with some questions, which are described as follows:
Here is my code
the output is
My questions are:
Many thanks!
(1) No, by definition tracers never appear or disappear.
In TNG50-1, at z=0, the minimum PartType3/TracerID is also 100000000002, so perhaps there is an issue in your loading code above. Could be subtle, i.e. related to memory? Since these are large arrays.
(2) It doesn't. Tracers are generated in the ICs (at t=0) only.
Hi! I re-run some similar codes in the TNG officical
JupterLab Workspace, and the complete code is as follows,the output of snapshot at z=0 is
I also run the similar one but for z=2
the output of snapshot at z=2.0 is
It is very confusing.
The figure of output
It's a bit complex, I would suggest a simpler test:
gives
Note: the min/max in any particular chunk is not important, only the global (across all chunks) is.
Got it! But the global minimum TracerID at z=2.0 and z=0 is still different. In the previous code, I run it in my local machine via load the tracers of the whole snapshot. In the latter code, I run it in TNG JupterLab where I only have a memory no more than 10GB and must split them in chunks. Both of them give a different global minmum TracerID at z=2.0 and z=0.
Hi! I think I locate the problem.
In the last two kinds of codes mentioned above, both of them use
il.snapshot.loadSubset. In the last code you gave, it usesh5pyto load the snapshot files. I also try to load the snapshot file viah5pydirectly and found the minimum and maximum values of TracerID across snapshots are the same.In short, do not use
il.snapshot.loadSubsetto load tracer particles and you had better to load it viah5py.Hi! I guess that I figured the reason:
il.snapshot.loadSubsetwill use the number,header['NumPart_Total'][PartTypeIndex['tracers']], which has a value of 1487761408. However, there is 10077696000 tracer particles. Such a larger number can also be found in the url TNG50-1 Data ana be obtained by adding up the numberheader['NumPart_ThisFile'][PartTypeIndex['tracers']]of the total 680 files (for TNG50-1).The corresponding codes are
It still confuses me why there are these two numbers.
Due to the limits on 32 bit numbers, slightly more work is needed to obtain total particle counts from the headers, as implemented in the getNumPart() function.
Got it! After some tests, I found that there may be a
bughere. Let me explain it step by step:The original code of
getNumPartisheader['NumPart_Total_HighWord']of TNG 50-1 isarray([2, 2, 0, 2, 0, 0], dtype=uint32)header['NumPart_Total']of TNG50-1 isarray([ 802737432, 1487761408, 0, 1487761408, 341411850, 3028], dtype=uint32)j=3ingetNumPart, and the total number of tracer particles isheader['NumPart_Total_HighWord'][3] *2**32 + header['NumPart_Total'][3], which is2^32 + 1487761408.getNumPart, the data type ofheader['NumPart_Total_HighWord'][3]isnp.uint32, soheader['NumPart_Total_HighWord'][3] *2**32(header['NumPart_Total_HighWord'][3] <<32exceeds the maximum limit ofnp.uint32, thenheader['NumPart_Total_HighWord'][3] <<32is0, the total number returned bygetNumPartis wrong.getNumParttoI found that the code in getNumPart function solved such a bug 10 months ago. I was provided the account of TNG JupyterLab about 3 years ago, so the package is of the older version.
Hi yes I see, that seems correct.
Unfortunately you're right - Lab accounts are given some starting files when they are created, and these are not automatically updated. This situation could be improved.
You can delete your
illustris_pythonand get the newest version from github.