Issue with Illustris tutorial: error fie not found
Sophia Nasr
1 Apr '19
Hello, was going through the Illustris script tutorial, and performed all the steps exactly as listed. I get an error on this line:
subhalos = il.groupcat.loadSubhalos(basePath,135,fields=fields)
It is as follows:
OSError: Unable to open file (unable to open file: name = './Illustris-3/output//groups_135/fof_subhalo_tab_135.1.hdf5', errno = 2, error message = 'No such file or directory', flags = 0, o_flags = 0)
So, I looked into the folder where the files that were downloaded using wget went (groups_135) and, of course, I found that the above file 'fof_subhalo_tab_135.1.hdf5' does not exist; only 'groupcat-135.1.hdf5' and 'groups_135.0.hdf5' are available. I also looked into the snapshot for Illustris 3 itself and found indeed only these two files exist. So my question is, is it possible that some naming scheme changed which causes the script listed to look for the wrong file, or am I supposed to download it separately?
Thanks.
Dylan Nelson
1 Apr '19
Hi Sophia,
If you look at the top of the groupcat.py file, you can see the search is quite simple. First it looks for something like groups_135.0.hdf5, and if this isn't found, then it looks for something like fof_subhalo_tab_135.0.hdf5.
It seems you are missing the second chunk (fof_subhalo_tab_135.1.hdf5) or else have the wrong path?
Sophia Nasr
1 Apr '19
Hi Dylan,
Okay I see that line in the code. *I think, then, that it wants the groups-135.1.hdf5 file, but the only file that's located in the folder with a 1 after the 135 is the groupcat file, groupcat-135.1.hdf5 (which I downloaded with wget). I did try downloading again, and got the file groupcat-135.0.hdf5, which was not there before, but for some reason no matter what I do, I cannot get the groups-135.1.hdf5 file! I looked directly into the snapshot by going to the Illustris-3 link (in the list of resolutions), and went own to snapshot 135, and in the group catalog the file doesn't exist. I tried using wget several times thnking maybe it skipped it, but it looks like that's not the case. Can I just download the specific file on my own??
it will download two files and name them groups_135.0.hdf5 and groups_135.1.hdf5.
Or you can just download by clicking on the two links.
If they have the wrong name for some reason, you can rename them.
Les Dickens
3 May '22
Hello, I am working through the Illustris script tutorial and getting almost exactly the same problem as described here, i.e. OSError: Unable to open file (unable to open file: name = './Illustris-3/output//groups_135/fof_subhalo_tab_135.1.hdf5'...etc.
In my case the 2 files, groups_135.0.hdf5 and groups_135.1.hdf5 are both in the downloaded catalogue; nevertheless the search at the top of groupcat.py still seems to fail and is looking for fof_subhalo_tab_135.1.hdf5 instead, which is not in the catalogue.
Looking through the script Traceback, the only thing I have found which seems related is that the variable snapNum, which is used in the search, is not defined. Should it be? Have I missed something?
Any ideas where to look next would be very welcome....
Thanks,
Les
Dylan Nelson
3 May '22
I suspect it is just a path issue, please double check the paths. If it is looking for the file './Illustris-3/output//groups_135/fof_subhalo_tab_135.1.hdf5' then this file, with that path, relative to the working directory of your python process, needs to exist.
Les Dickens
3 May '22
Hi, thanks for the response. I checked the path and it does exist. Just to be sure I changed the directory name from Illustris-3 to “IllustrisNEW” and also changed the “basePath=...” command to use the new name. Sure enough, the same OSError came back refencing the changed name.
The directory (~/Illustris-3/output/groups_135/) contains the two files: groups_135.0.hdf5 and groups_135.1.hdf5 and nothing else. You pointed out above, that groupcat.py first looks for something like groups_135.0.hdf5, and if this isn't found, then it looks for something like fof_subhalo_tab_135.0.hdf5. I did look at the code, but I’m a novice – I’ll take your word for it :). Is there anything else I could look at, to try and figure out what’s going wrong?
Thanks again,
Les
Dylan Nelson
3 May '22
Please post a complete example (all commands and outputs)?
Les Dickens
4 May '22
Path error - you were correct. Fixed it now.
Thanks,
Les
Pradyumna Sadhu
2 Jan '23
@Les, what change did you make? I am still unable to figure this out
@Pradyumna
This may not help much, but in my case it was a simple "typo". The leading "." was missing in the statement : basePath = './Illustris-3/output/; that caused the error. I hope that's some help....
Thanks you for the reply! However, here is what worked for me:
In the gcPath function of groupcat.py, I had to change the following two lines: gcPath = basePath + '/groups_%03d/' % snapNum filePath1 = gcPath + 'groups_%03d.%d.hdf5' % (snapNum, chunkNum)
to gcPath = basePath + '/groups_%13d/' % snapNum filePath1 = gcPath + 'groups_%13d.%d.hdf5' % (snapNum, chunkNum)
since the files were downloaded as groups_135.0.hdf5 and so on, but not groups_035.0.hdf5
Thắng Huỳnh
4 Aug '23
Hi everyone,
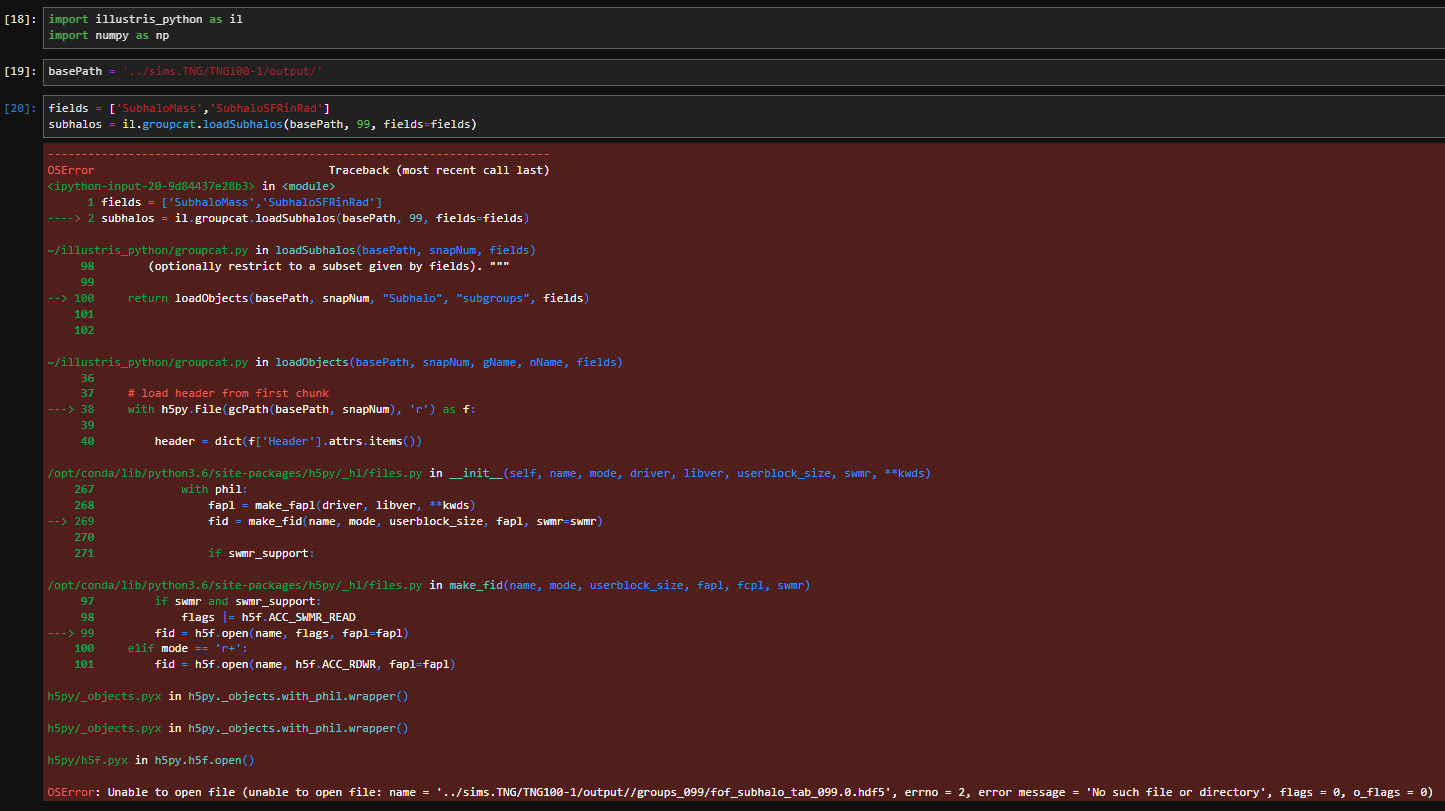
I am having the similar circumstance now albeit a little different. In my case, when I use the file tutorial.ipynb in the examples path, the cell: fields = ['SubhaloMass','SubhaloSFRinRad'] subhalos = il.groupcat.loadSubhalos(basePath, 99, fields=fields)
works fine.
However, when I try to create a new notebook and run the same code, it returns this error:
OSError: Unable to open file (unable to open file: name = '../sims.TNG/TNG100-1/output//groups_099/fof_subhalo_tab_099.0.hdf5', errno = 2, error message = 'No such file or directory', flags = 0, o_flags = 0)
Thank you in advance,
Thang
Dylan Nelson
4 Aug '23
This is just a path issue. Where is the "new notebook"? Make sure you save it.
If you save it directly into your home directory, then you would want a path of sims.TNG/TNG100-1/....
If you save it into the examples directory, or any sub-directory, you would need to have a path of ../sims.TNG/TNG100-1/...
Thắng Huỳnh
4 Aug '23
I saved it in the examples directory and to make sure, I tried to copy the path from the "tutorial.ipynb" code but it still returned error.
By the way, I still don't know where is called home directory.
Dylan Nelson
4 Aug '23
I would suggest googling for a introductory linux tutorial on paths and the command line. Then you can verify about the home directory, the examples directory, and the "relative path" you are using.
Thắng Huỳnh
4 Aug '23
Sorry but I am using the web-based Lab session, so it is matter?
Hello, was going through the Illustris script tutorial, and performed all the steps exactly as listed. I get an error on this line:
subhalos = il.groupcat.loadSubhalos(basePath,135,fields=fields)
It is as follows:
OSError: Unable to open file (unable to open file: name = './Illustris-3/output//groups_135/fof_subhalo_tab_135.1.hdf5', errno = 2, error message = 'No such file or directory', flags = 0, o_flags = 0)
So, I looked into the folder where the files that were downloaded using wget went (groups_135) and, of course, I found that the above file 'fof_subhalo_tab_135.1.hdf5' does not exist; only 'groupcat-135.1.hdf5' and 'groups_135.0.hdf5' are available. I also looked into the snapshot for Illustris 3 itself and found indeed only these two files exist. So my question is, is it possible that some naming scheme changed which causes the script listed to look for the wrong file, or am I supposed to download it separately?
Thanks.
Hi Sophia,
If you look at the top of the groupcat.py file, you can see the search is quite simple. First it looks for something like
groups_135.0.hdf5, and if this isn't found, then it looks for something likefof_subhalo_tab_135.0.hdf5.It seems you are missing the second chunk (
fof_subhalo_tab_135.1.hdf5) or else have the wrong path?Hi Dylan,
Okay I see that line in the code. *I think, then, that it wants the groups-135.1.hdf5 file, but the only file that's located in the folder with a 1 after the 135 is the groupcat file, groupcat-135.1.hdf5 (which I downloaded with wget). I did try downloading again, and got the file groupcat-135.0.hdf5, which was not there before, but for some reason no matter what I do, I cannot get the groups-135.1.hdf5 file! I looked directly into the snapshot by going to the Illustris-3 link (in the list of resolutions), and went own to snapshot 135, and in the group catalog the file doesn't exist. I tried using wget several times thnking maybe it skipped it, but it looks like that's not the case. Can I just download the specific file on my own??
Hi Sophia,
If you run the wget command in the tutorial:
it will download two files and name them
groups_135.0.hdf5andgroups_135.1.hdf5.Or you can just download by clicking on the two links.
If they have the wrong name for some reason, you can rename them.
Hello, I am working through the Illustris script tutorial and getting almost exactly the same problem as described here, i.e. OSError: Unable to open file (unable to open file: name = './Illustris-3/output//groups_135/fof_subhalo_tab_135.1.hdf5'...etc.
In my case the 2 files, groups_135.0.hdf5 and groups_135.1.hdf5 are both in the downloaded catalogue; nevertheless the search at the top of groupcat.py still seems to fail and is looking for fof_subhalo_tab_135.1.hdf5 instead, which is not in the catalogue.
Looking through the script Traceback, the only thing I have found which seems related is that the variable snapNum, which is used in the search, is not defined. Should it be? Have I missed something?
Any ideas where to look next would be very welcome....
Thanks,
Les
I suspect it is just a path issue, please double check the paths. If it is looking for the file './Illustris-3/output//groups_135/fof_subhalo_tab_135.1.hdf5' then this file, with that path, relative to the working directory of your python process, needs to exist.
Hi, thanks for the response. I checked the path and it does exist. Just to be sure I changed the directory name from Illustris-3 to “IllustrisNEW” and also changed the “basePath=...” command to use the new name. Sure enough, the same OSError came back refencing the changed name.
The directory (~/Illustris-3/output/groups_135/) contains the two files: groups_135.0.hdf5 and groups_135.1.hdf5 and nothing else. You pointed out above, that groupcat.py first looks for something like groups_135.0.hdf5, and if this isn't found, then it looks for something like fof_subhalo_tab_135.0.hdf5. I did look at the code, but I’m a novice – I’ll take your word for it :). Is there anything else I could look at, to try and figure out what’s going wrong?
Thanks again,
Les
Please post a complete example (all commands and outputs)?
Path error - you were correct. Fixed it now.
Thanks,
Les
@Les, what change did you make? I am still unable to figure this out
Thanks in advance,
Pradyumna
@Pradyumna
This may not help much, but in my case it was a simple "typo". The leading "." was missing in the statement : basePath = './Illustris-3/output/; that caused the error. I hope that's some help....
Regards,
Les
Thanks you for the reply! However, here is what worked for me:
In the gcPath function of groupcat.py, I had to change the following two lines:
gcPath = basePath + '/groups_%03d/' % snapNumfilePath1 = gcPath + 'groups_%03d.%d.hdf5' % (snapNum, chunkNum)to
gcPath = basePath + '/groups_%13d/' % snapNumfilePath1 = gcPath + 'groups_%13d.%d.hdf5' % (snapNum, chunkNum)since the files were downloaded as groups_135.0.hdf5 and so on, but not groups_035.0.hdf5
Hi everyone,
I am having the similar circumstance now albeit a little different. In my case, when I use the file
tutorial.ipynbin theexamplespath, the cell:fields = ['SubhaloMass','SubhaloSFRinRad']subhalos = il.groupcat.loadSubhalos(basePath, 99, fields=fields)works fine.
However, when I try to create a new notebook and run the same code, it returns this error:
OSError: Unable to open file (unable to open file: name = '../sims.TNG/TNG100-1/output//groups_099/fof_subhalo_tab_099.0.hdf5', errno = 2, error message = 'No such file or directory', flags = 0, o_flags = 0)
Thank you in advance,
Thang
This is just a path issue. Where is the "new notebook"? Make sure you save it.
If you save it directly into your home directory, then you would want a path of
sims.TNG/TNG100-1/....If you save it into the examples directory, or any sub-directory, you would need to have a path of
../sims.TNG/TNG100-1/...I saved it in the examples directory and to make sure, I tried to copy the path from the "tutorial.ipynb" code but it still returned error.
By the way, I still don't know where is called home directory.
I would suggest googling for a introductory linux tutorial on paths and the command line. Then you can verify about the home directory, the examples directory, and the "relative path" you are using.
Sorry but I am using the web-based Lab session, so it is matter?
Nevermind, I don't know what happened, it just works now (I still use the web-based Lab).
Thank you for your time,
Thang